Learning objective
Summarise the experiments performed by the following scientists that provided evidence that DNA is the genetic material:
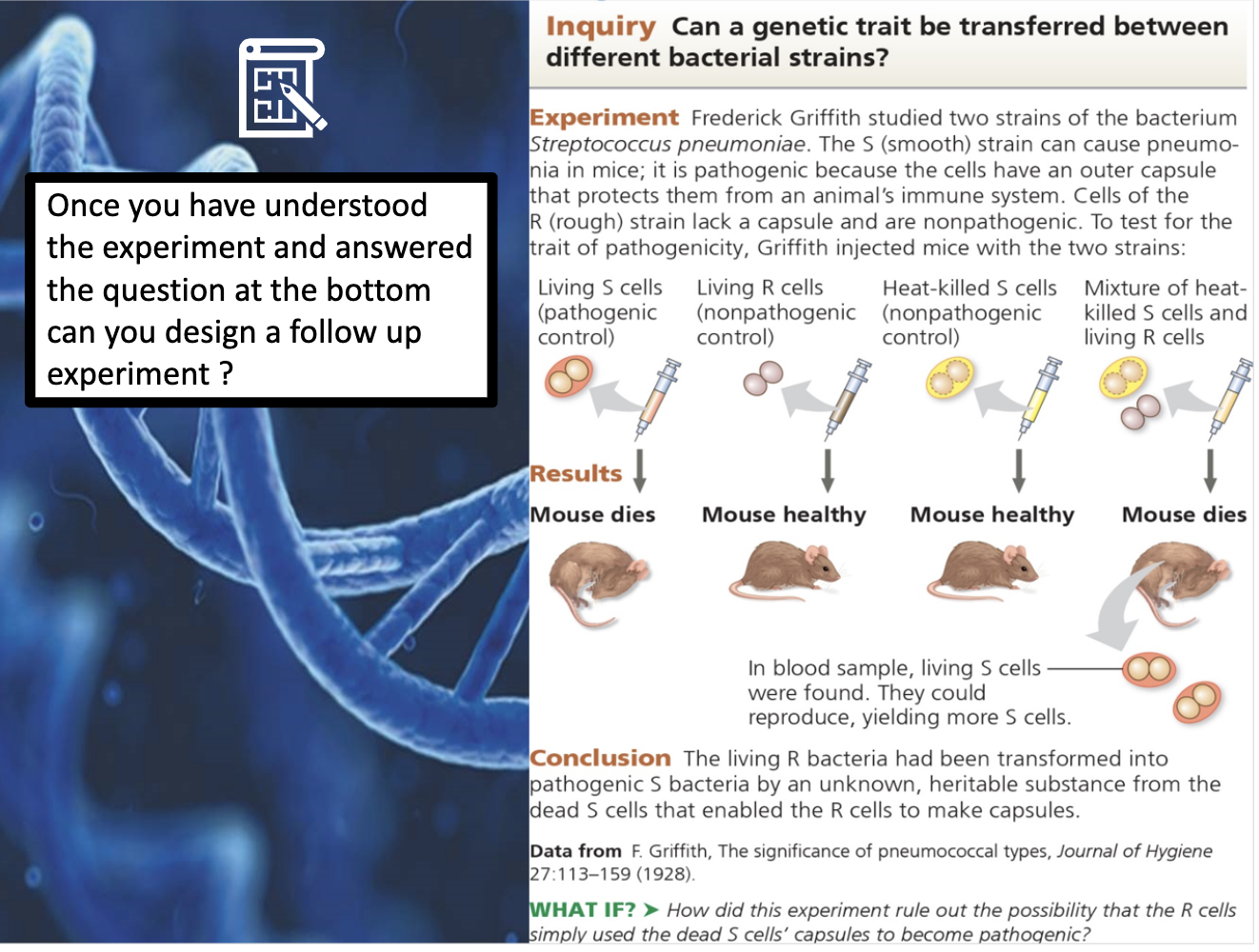
a. Frederick Griffith
By discovering substance in pathogenic cell transmitted to the other cell, he discovered there is an inheritance substance. He named the phenomenom he observed as transformation
b. Oswald Avery, Maclyn McCarty, and Colin MacLeod
They anounced that the transmitted substance is DNA
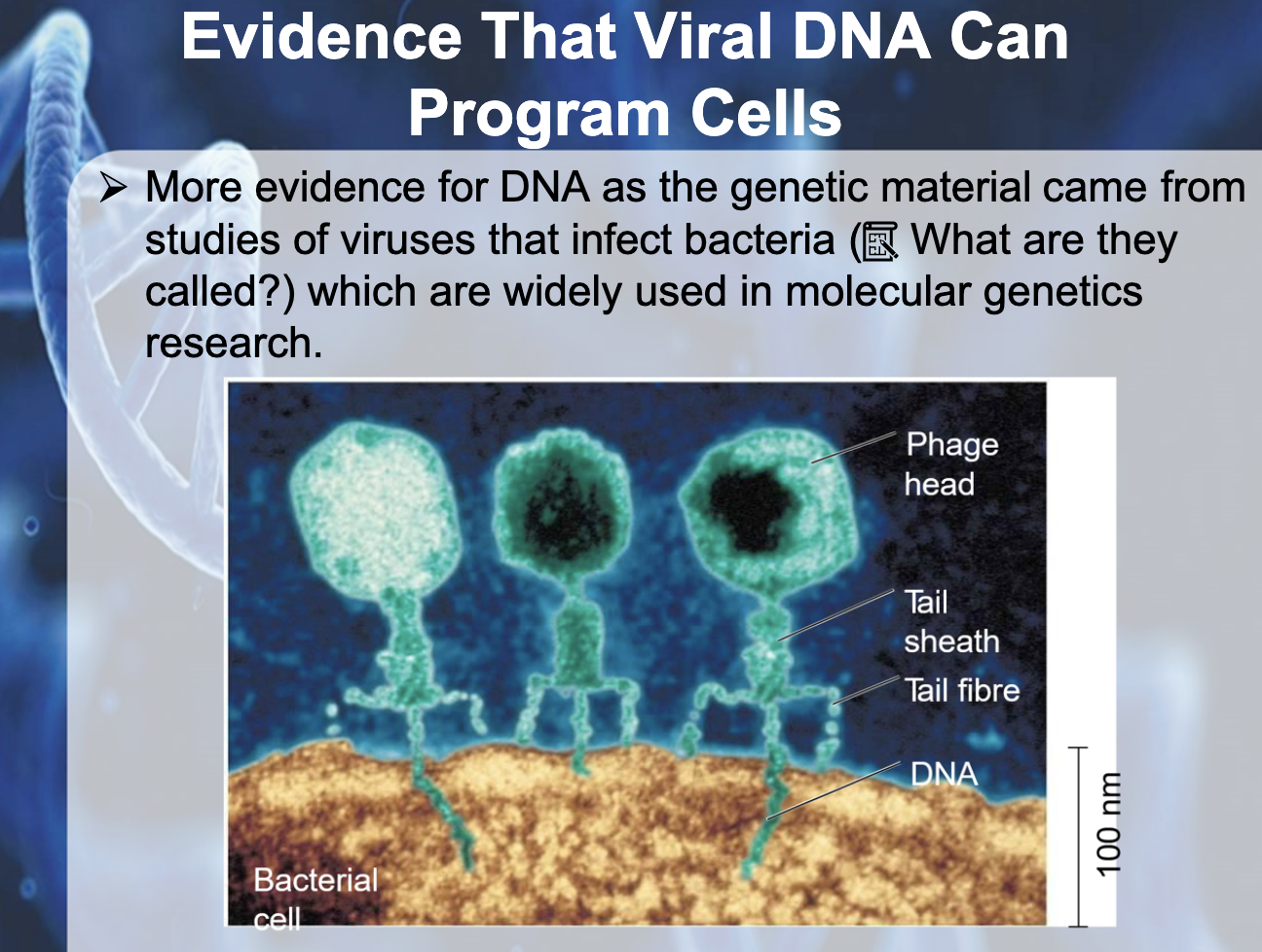
c. Alfred Hershey and Martha Chase
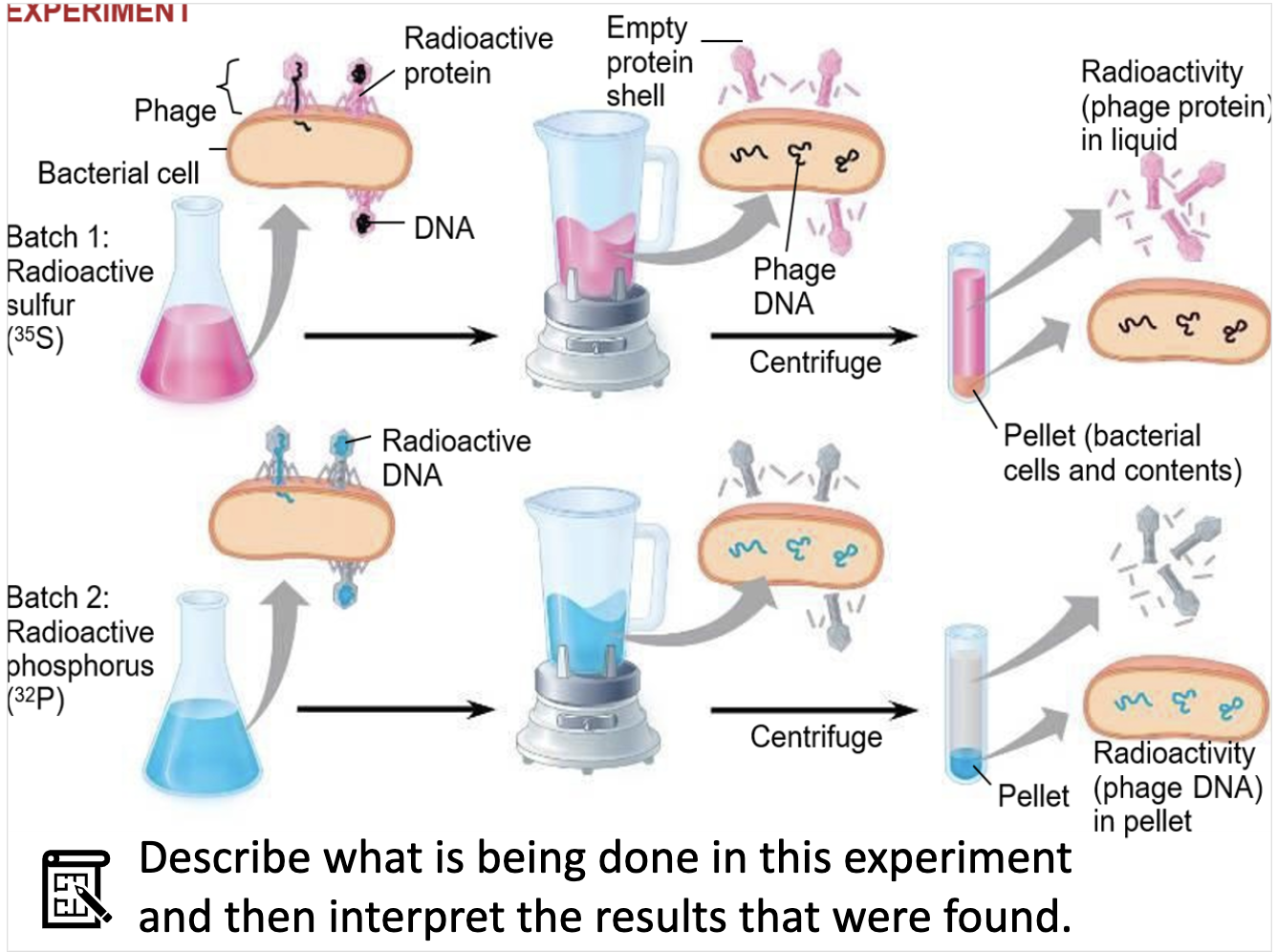
They had an experiment of discovering which part of the cell is transmitted, DNA or protein.
They used radioactive fluorescent that dyes Protien and DNA each.
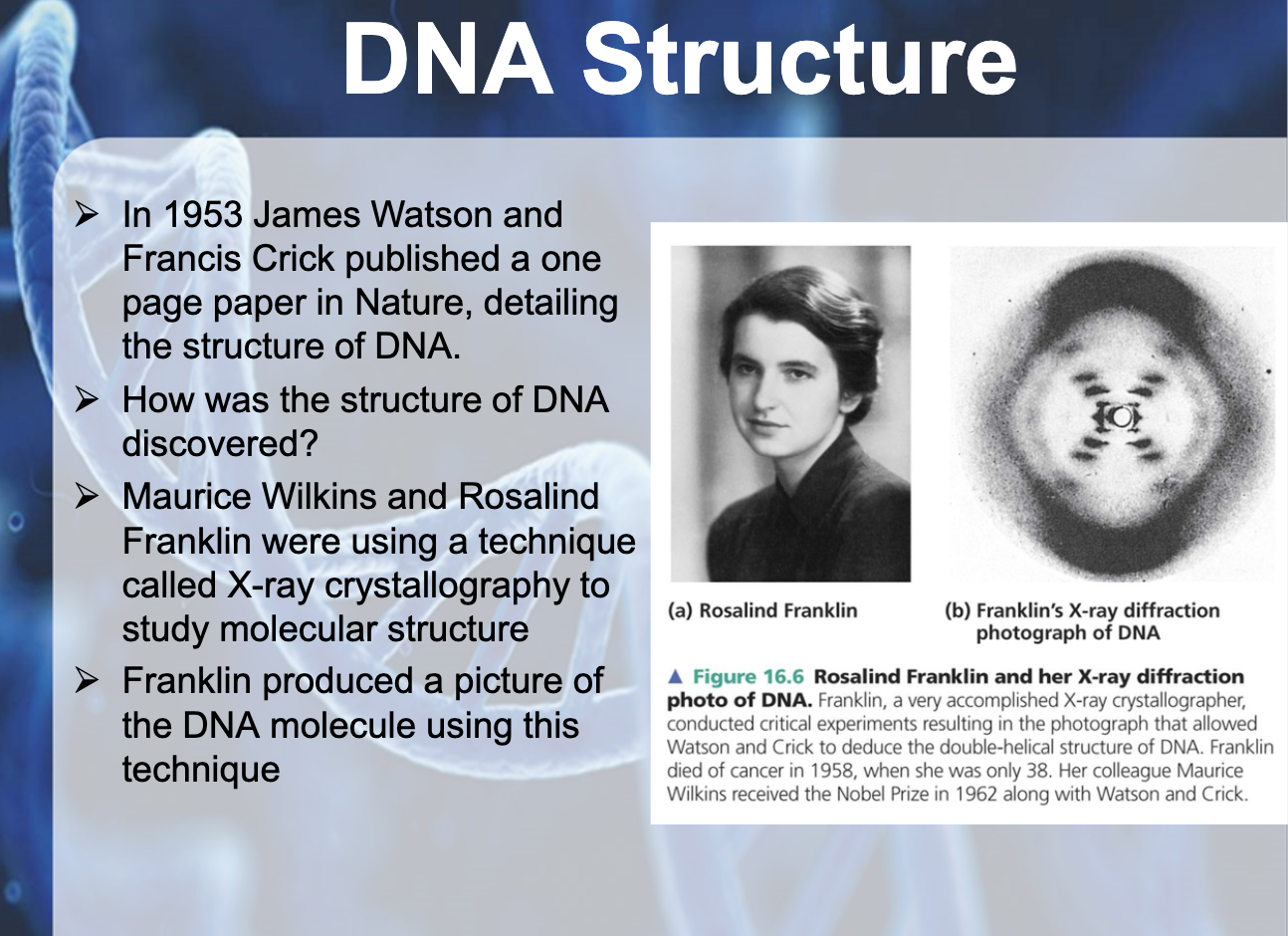
Explain the significance of the research of Rosalind Franklin, Maurice Wilkins, James Watson and Francis Crick in deducing the structure of DNA.
Rosalind Franklin and Maurice Wilkins developed X ray experiment to help James Watson and Fancis Crick to discover the structure of DNA and how they replicate them
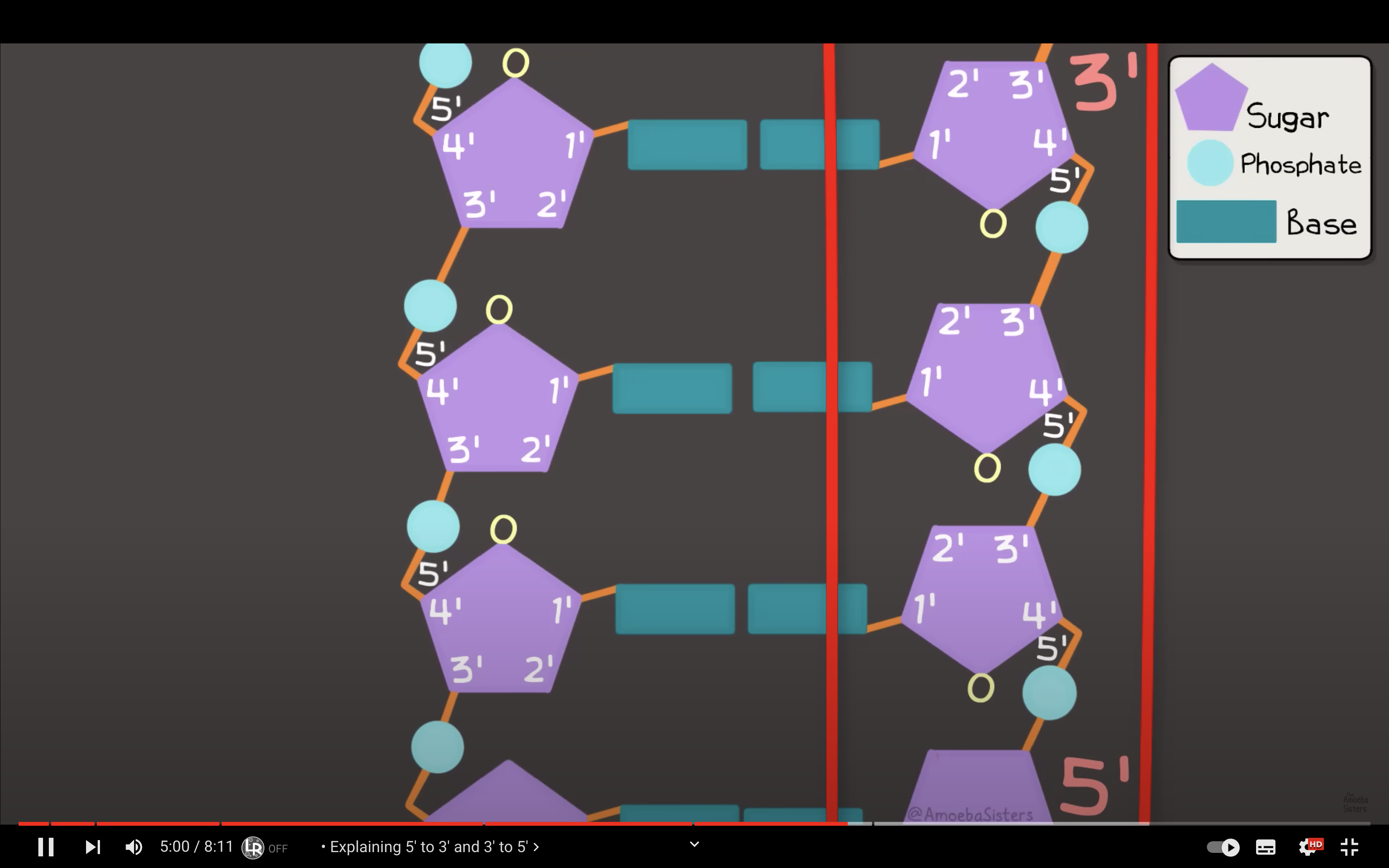
Describe the structure of DNA.
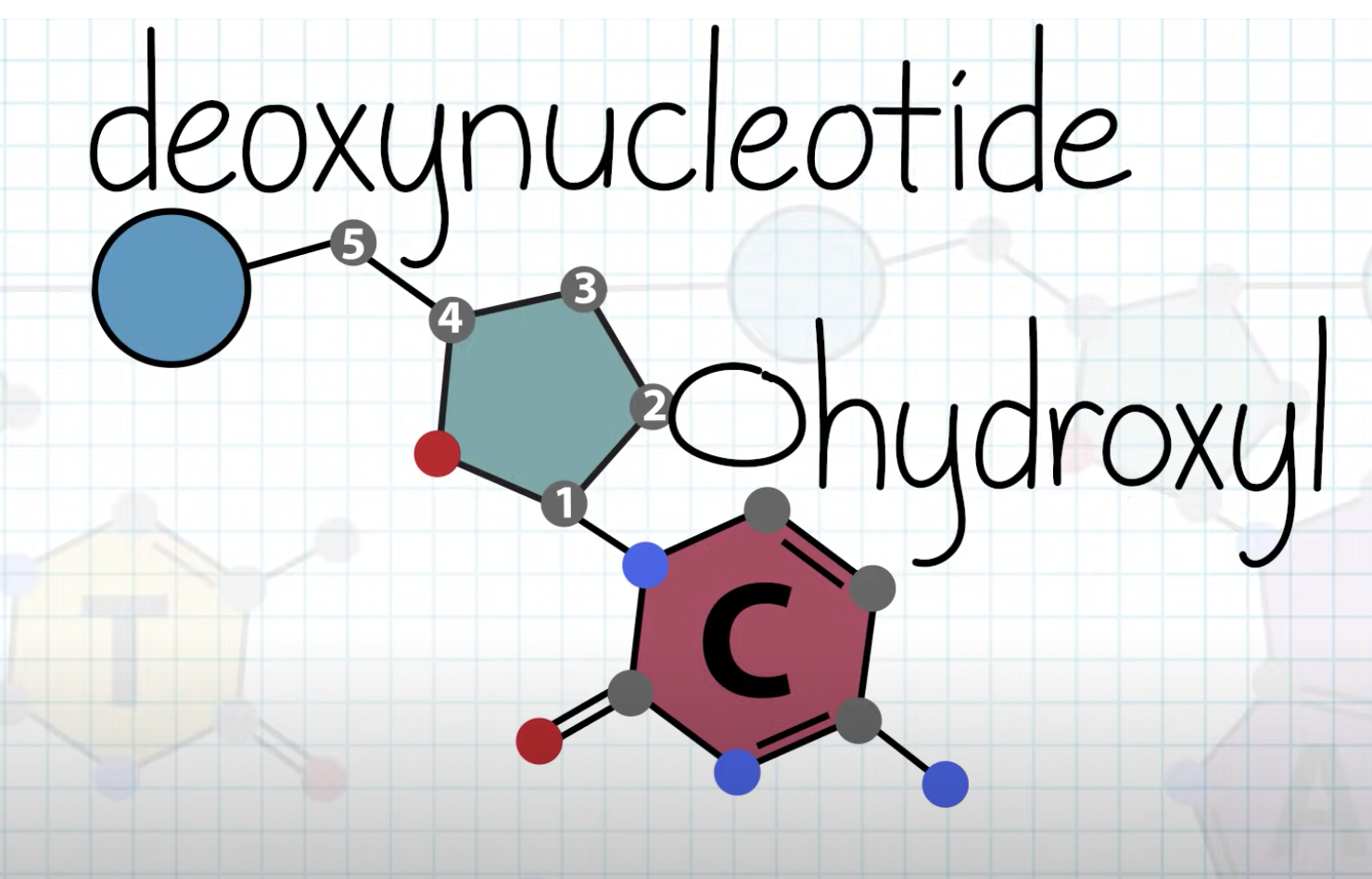
-deoxyribose + base + phosphate
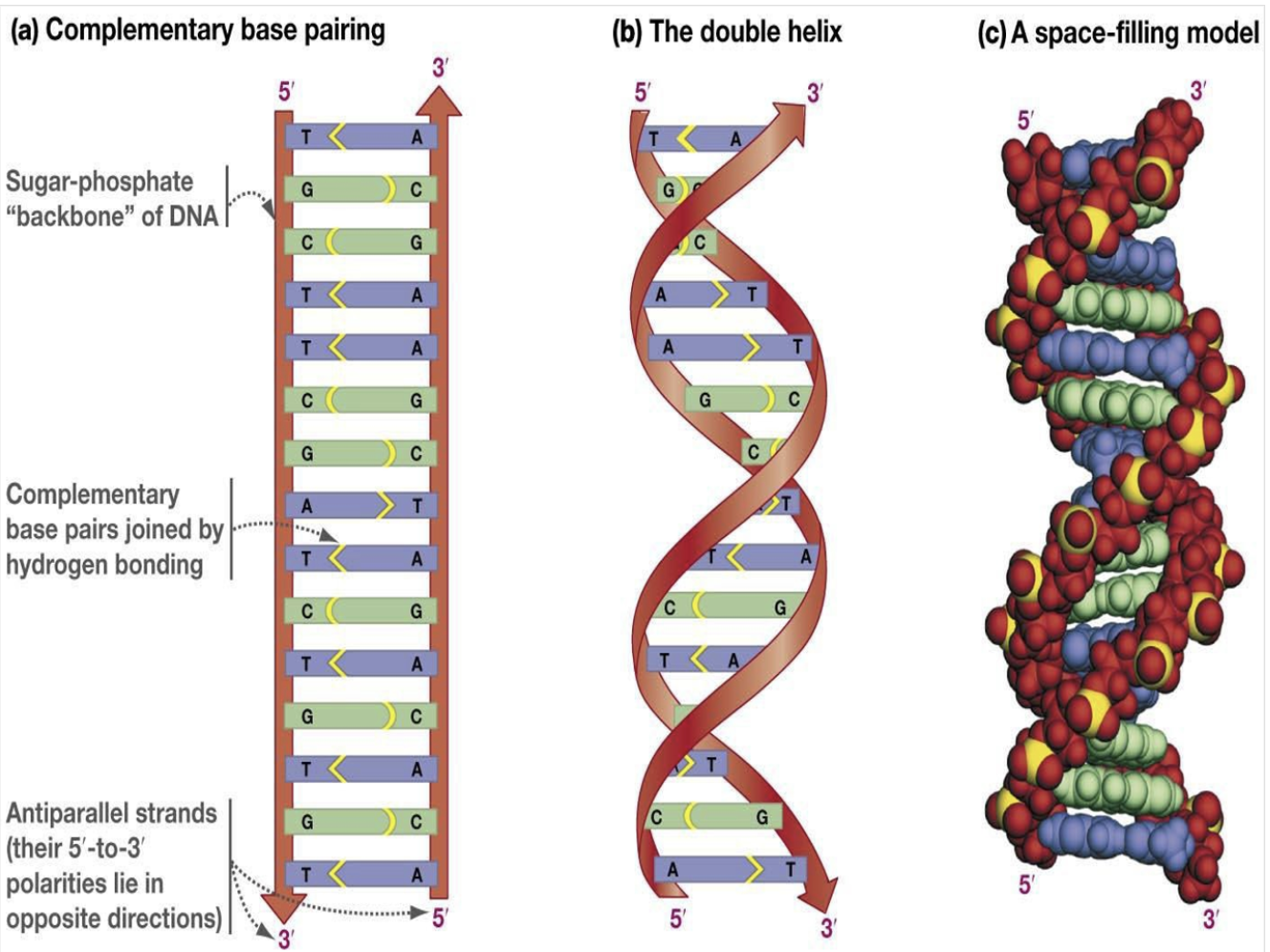
deoxyribose and phosphate consist the back bone of the DNA, and the base form hydrogen bonding with the complementary base of the other strand of the DNA.
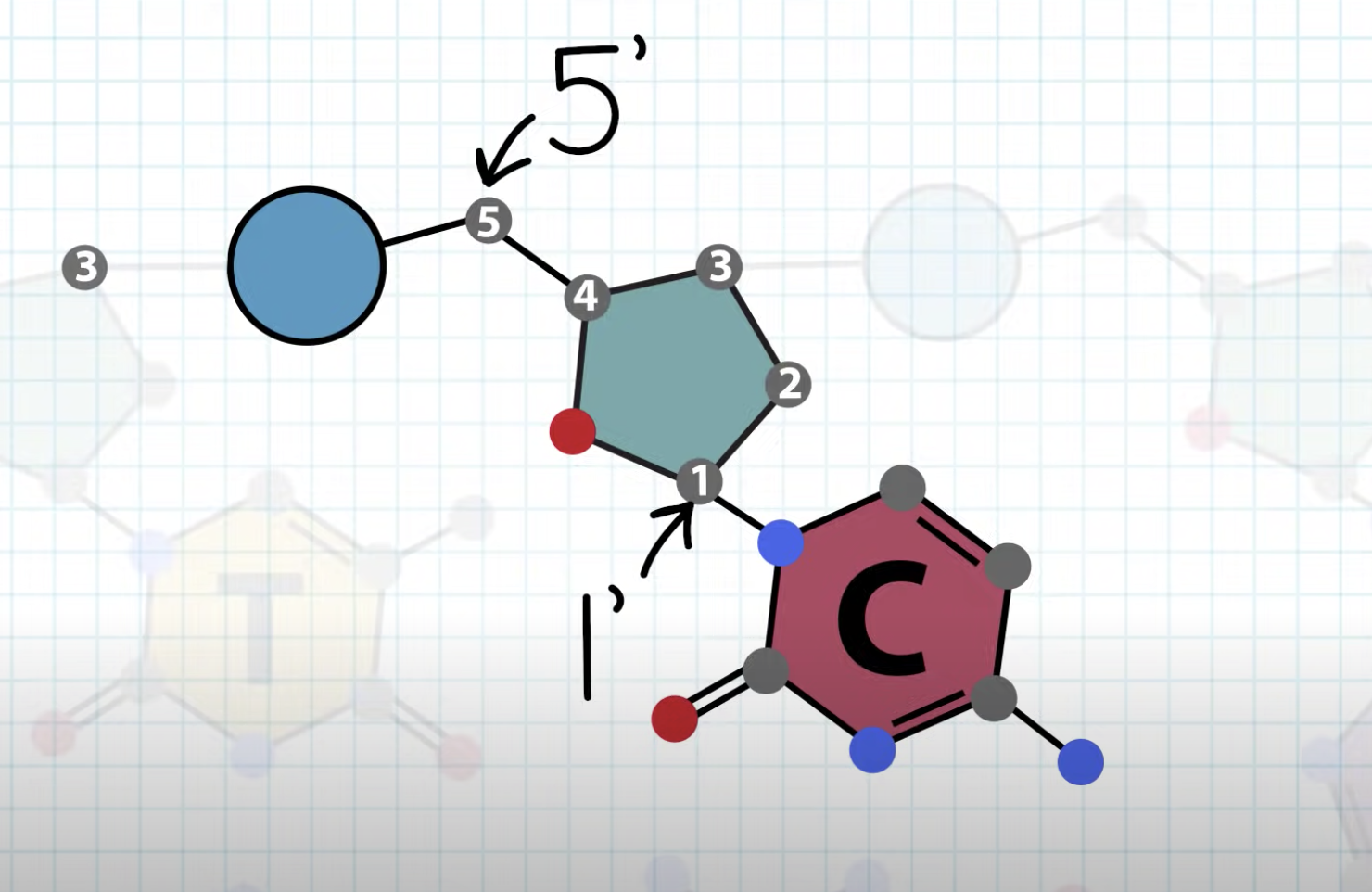
one strand is 5'->3' direction and the other strand is 3'->5' direction
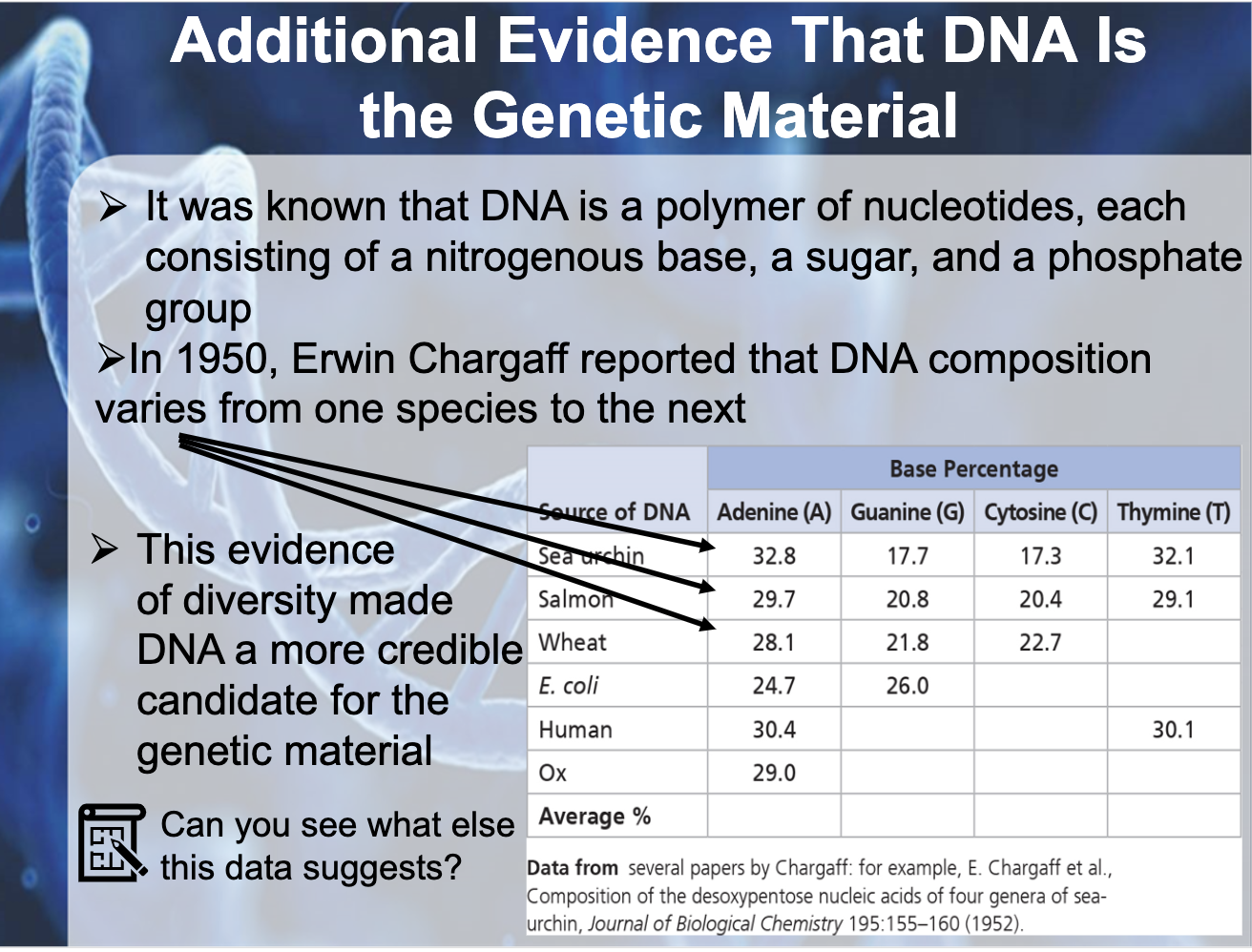
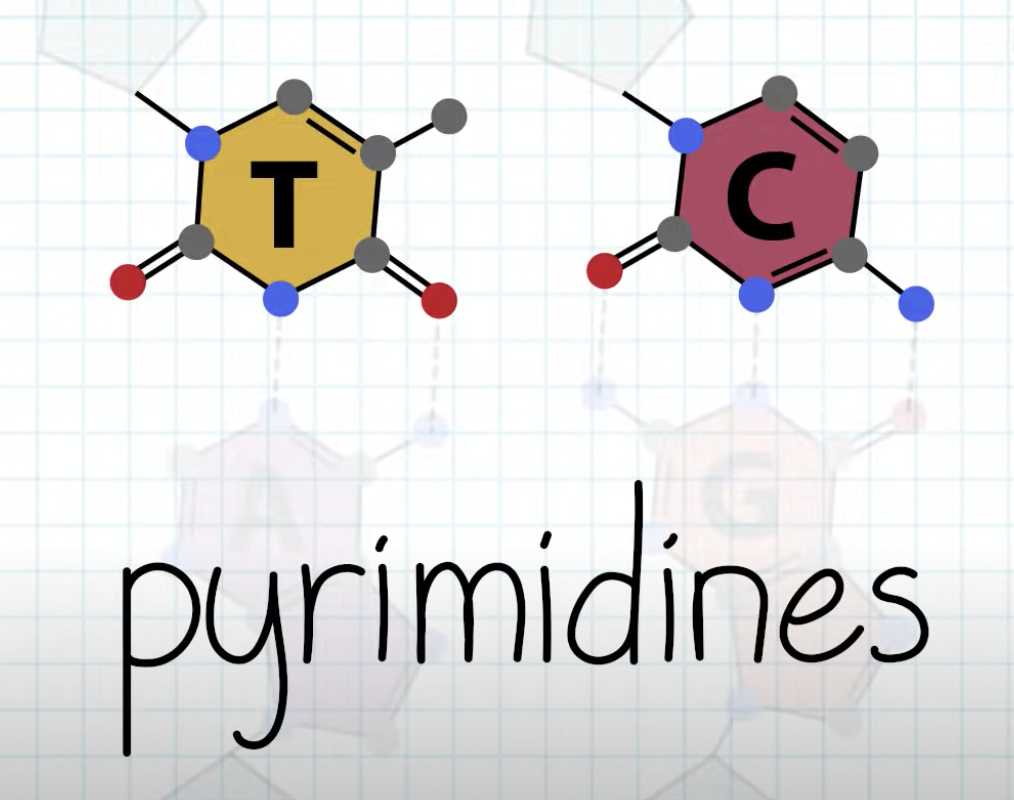
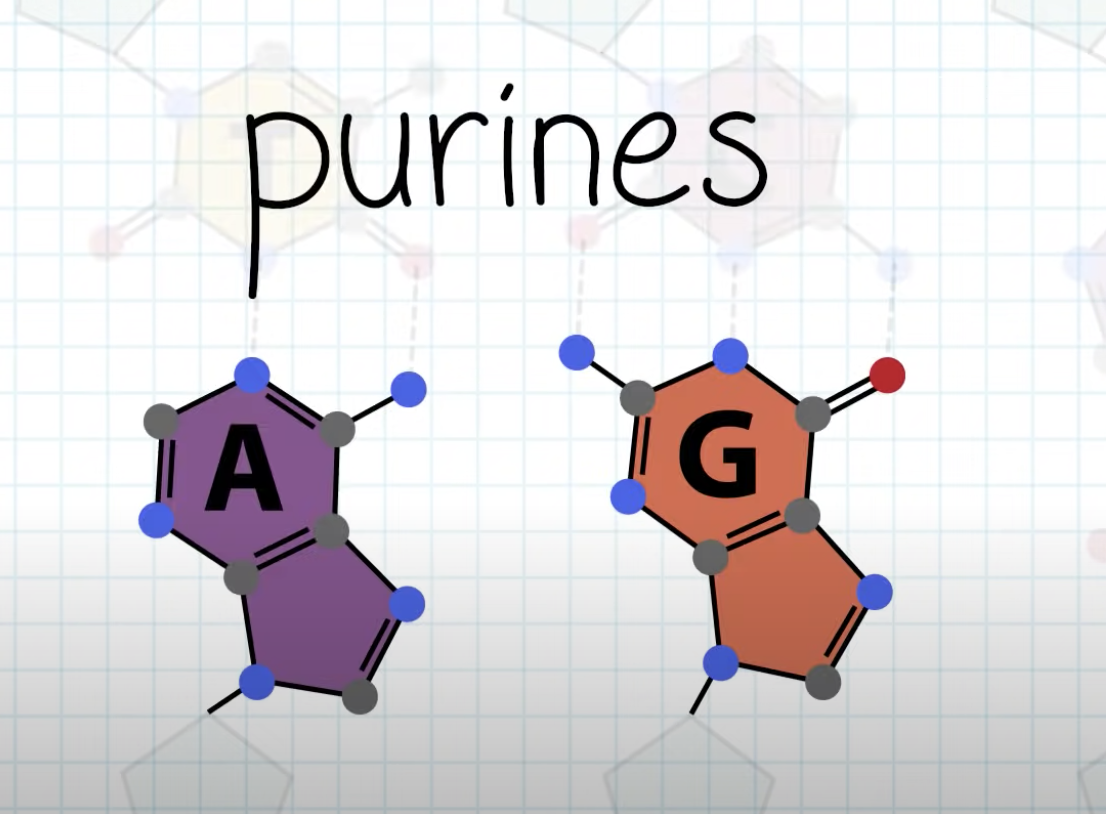
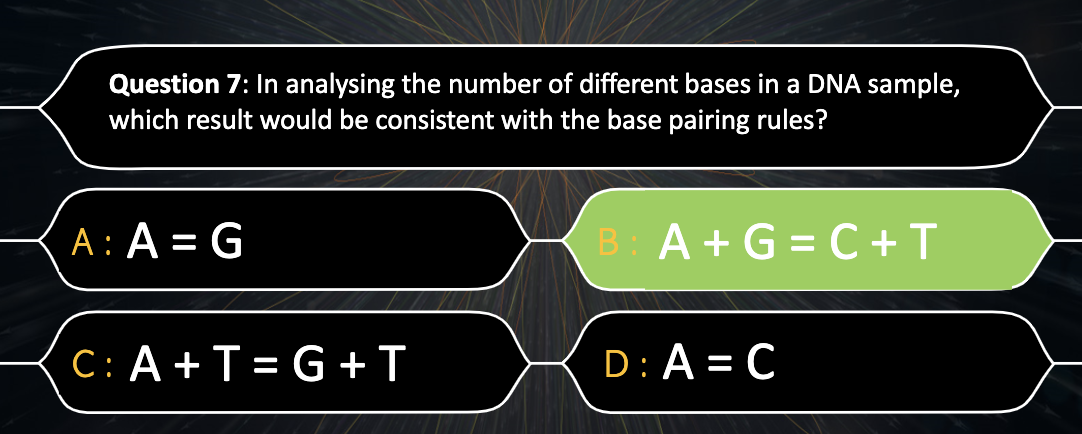
Explain Chargaff’s base-pairing rule and describe its significance.
A,G purine and T,C form consistent distance between the bonding.
Describe the semi-conservative model of replication and outline the experiment that confirmed this model as the correct model of DNA replication.
Each of DNA strands seperate to replicate. After the replication, two hybrid DNAs are produced. If they were expecting conservatve DNA replication model, after the replication, original strands should bind together again and result in whole new DNA double helix and complete original DNA double helix.
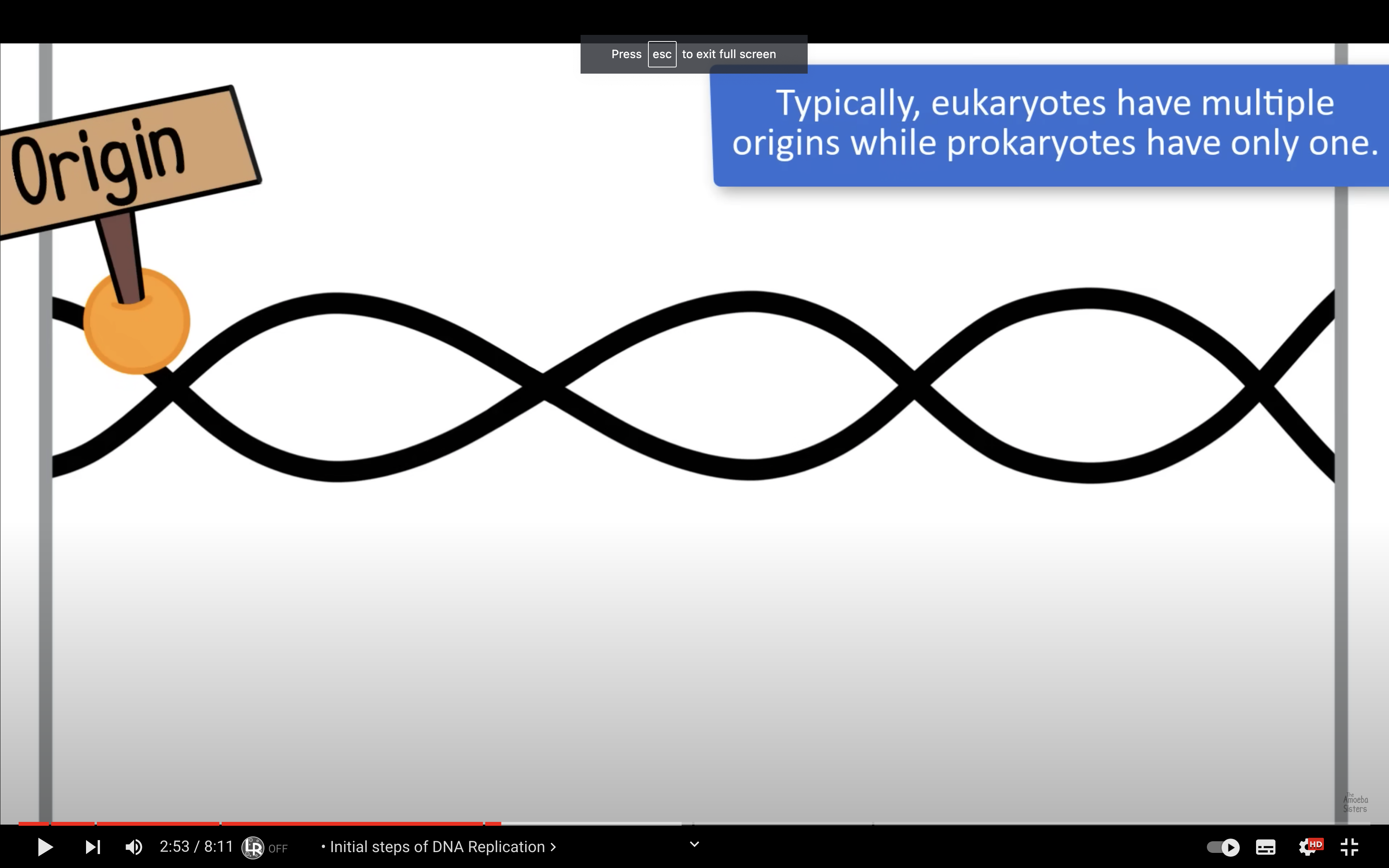
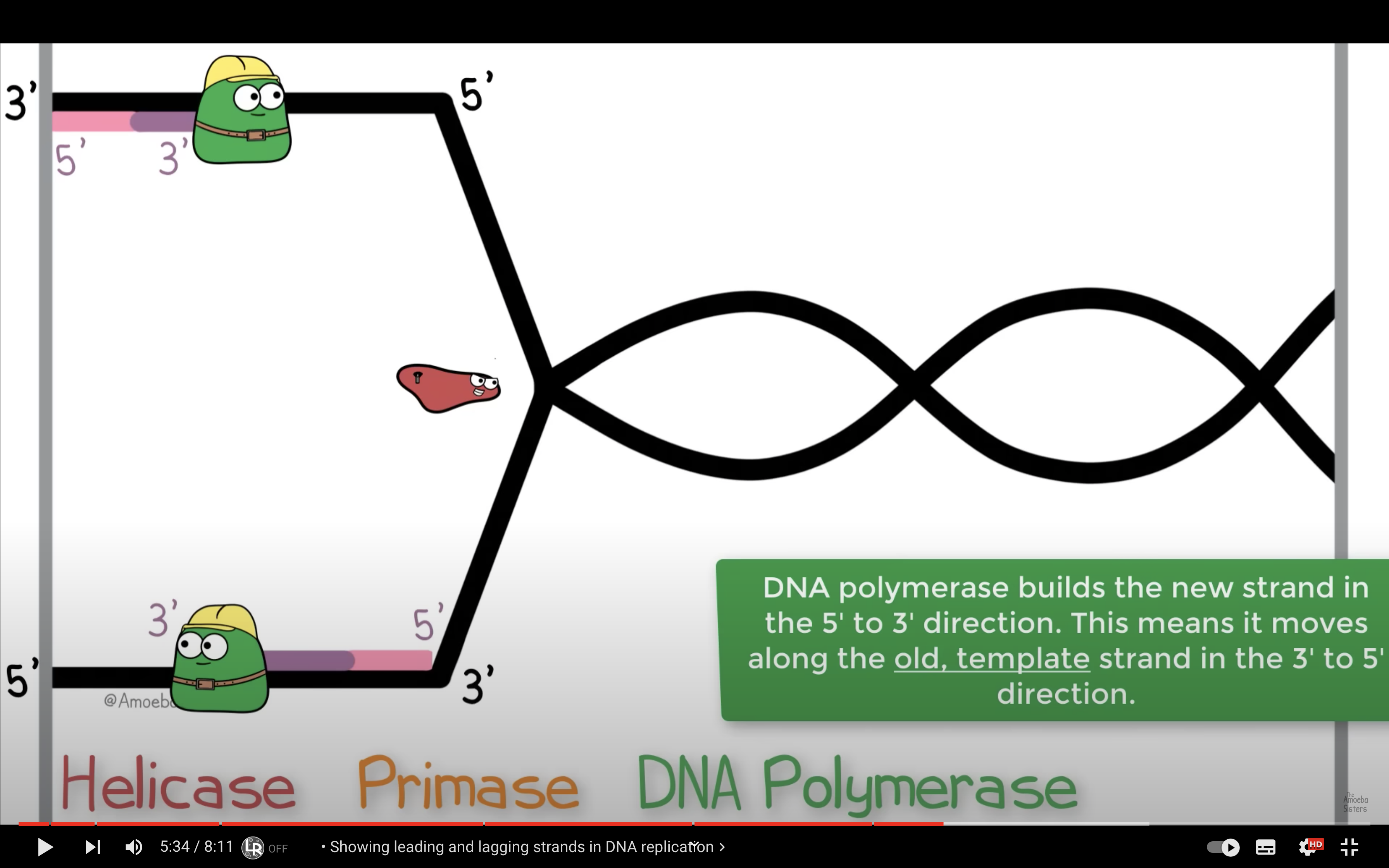
Describe the process of DNA replication, including the role of the replication fork and origin of replication.
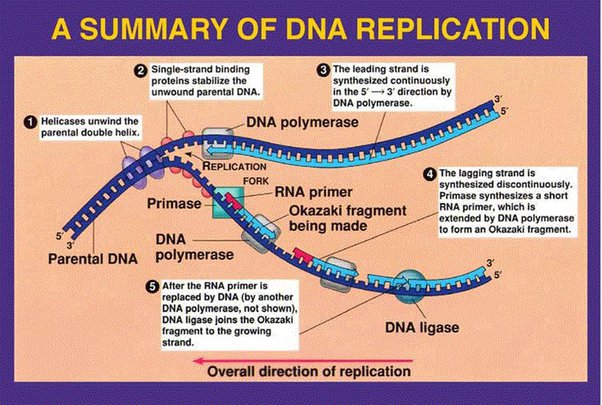
Origin of replication is the site where double strands seperate to form a bubble. Replication fork is the end of the bubble, Y-shaped region where helicase works to break H bonding between the double strands.
Occurs during s phase, replication begins at sites called origins of replication, where 2 DNA strands are separated and open up a replication 'bubble' with 2 replication forks. Enzymes are the involved which leads to a bi-directional synthesis; both leading and lagging strand are copied at the same time.
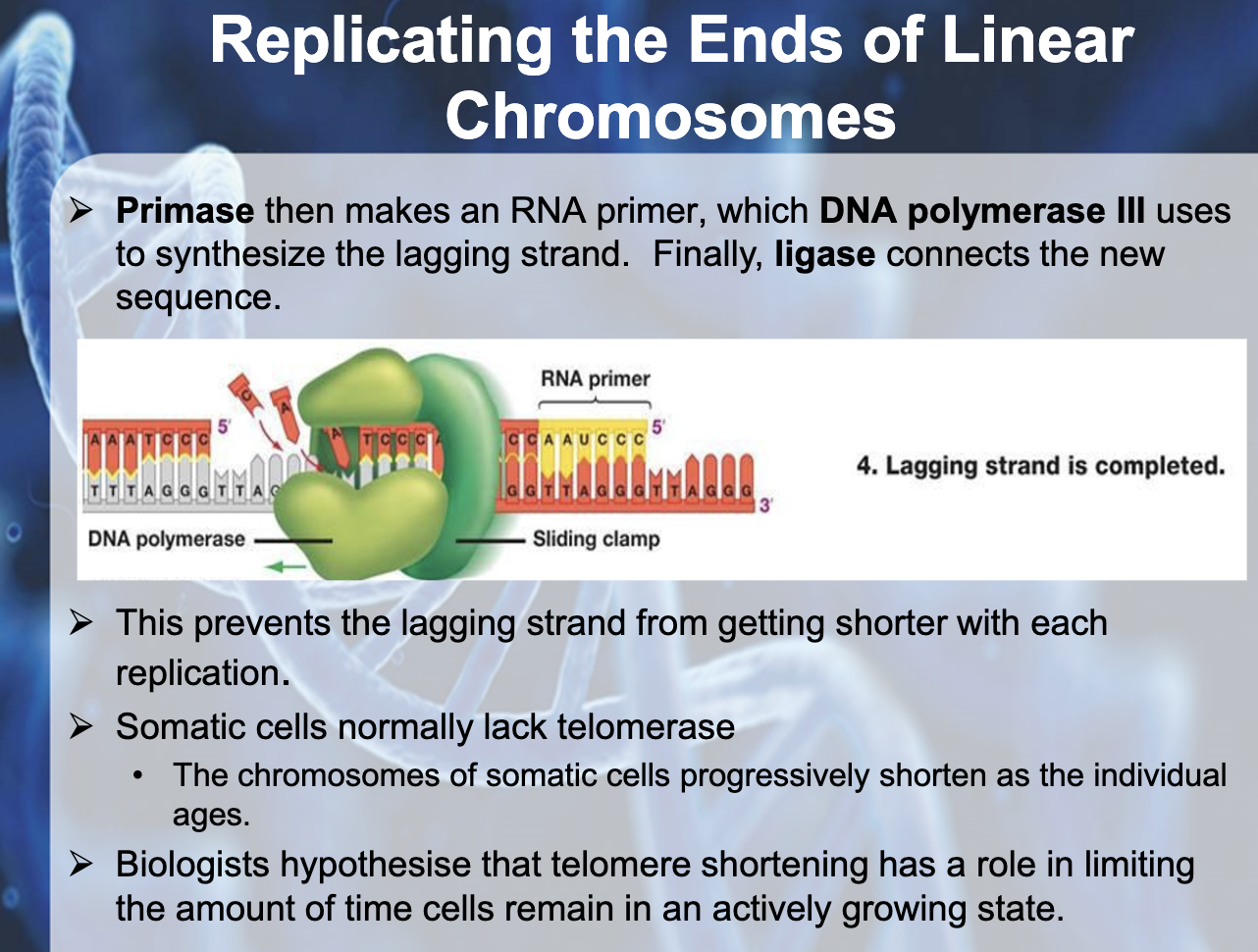
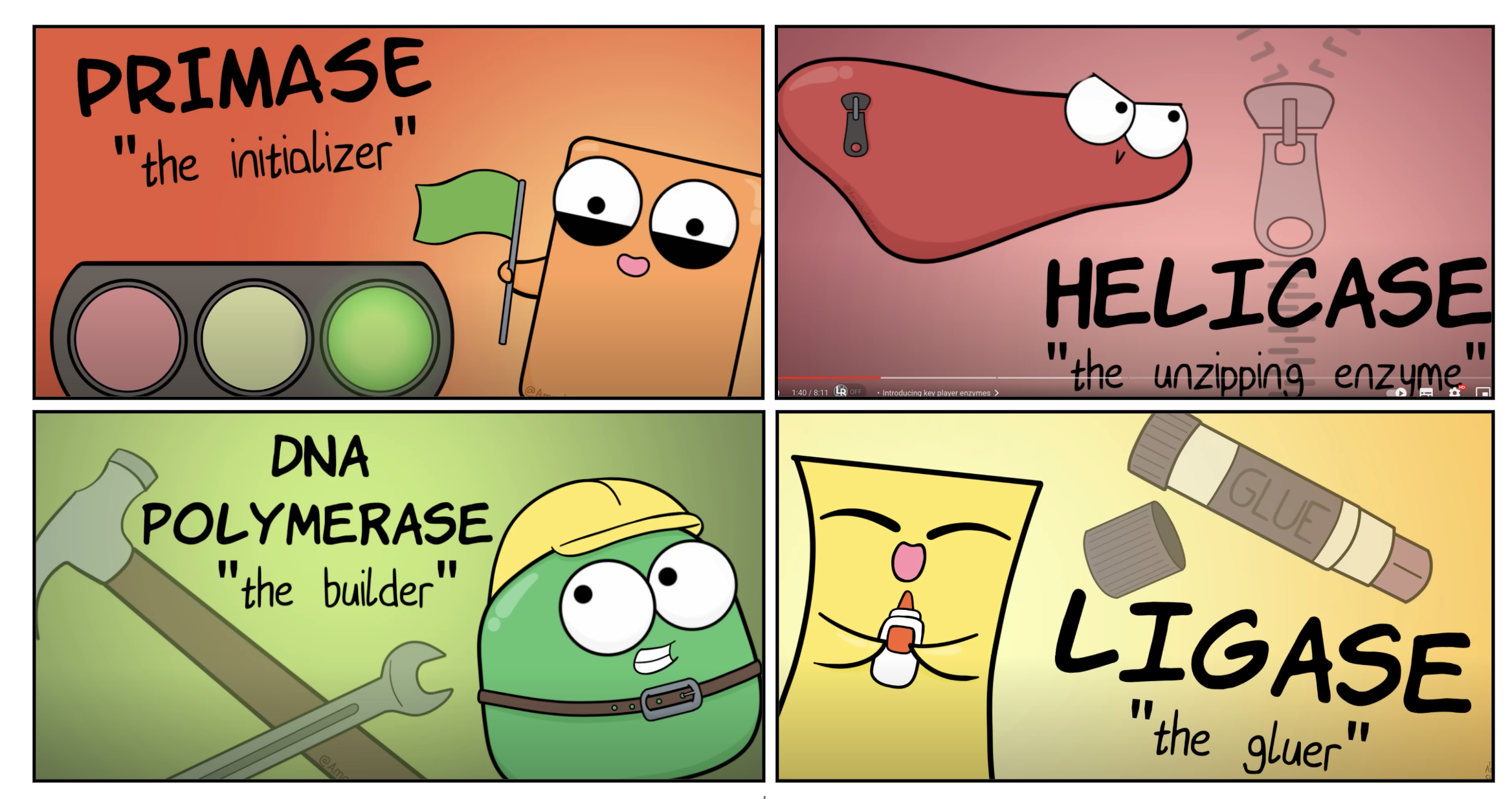
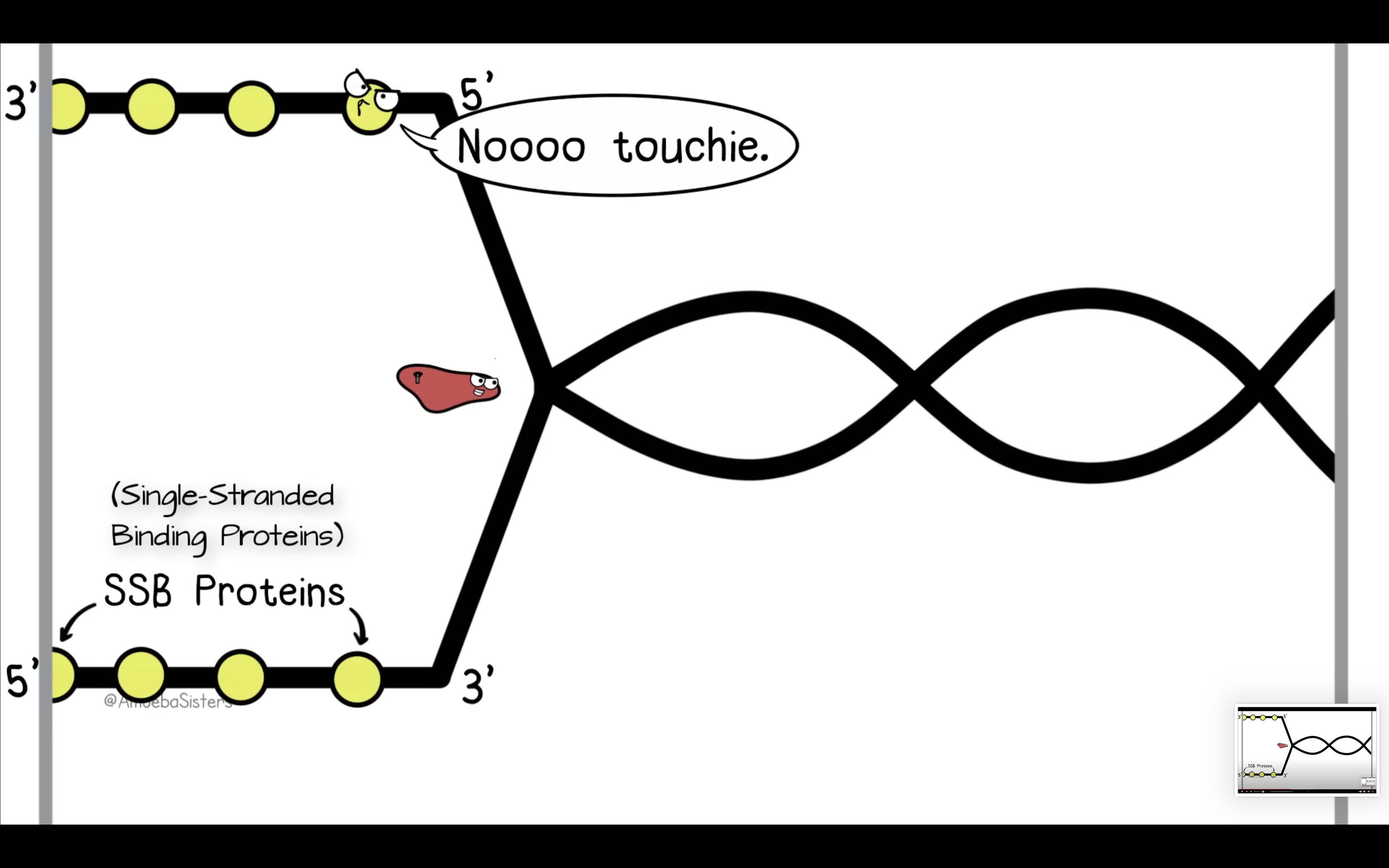
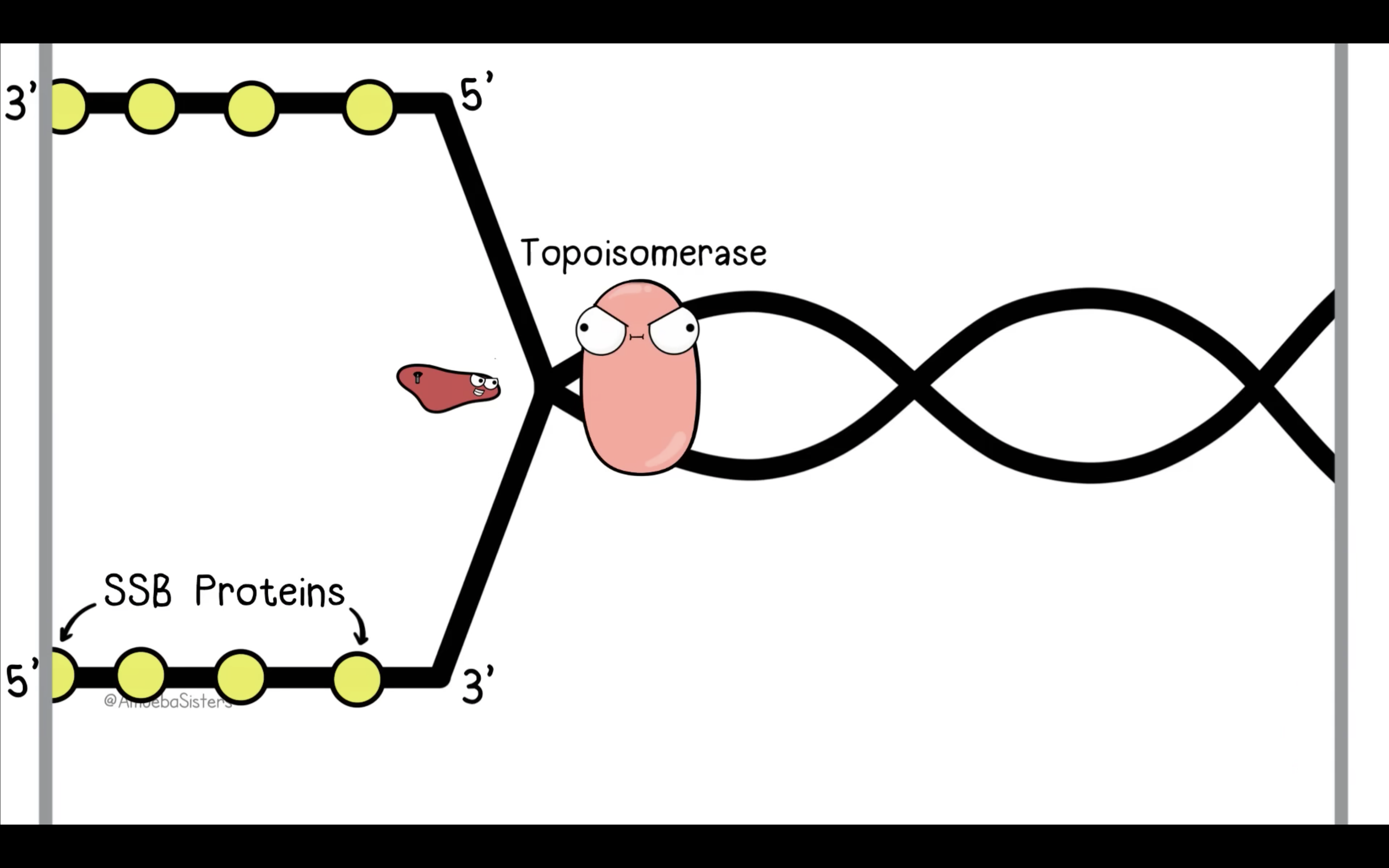
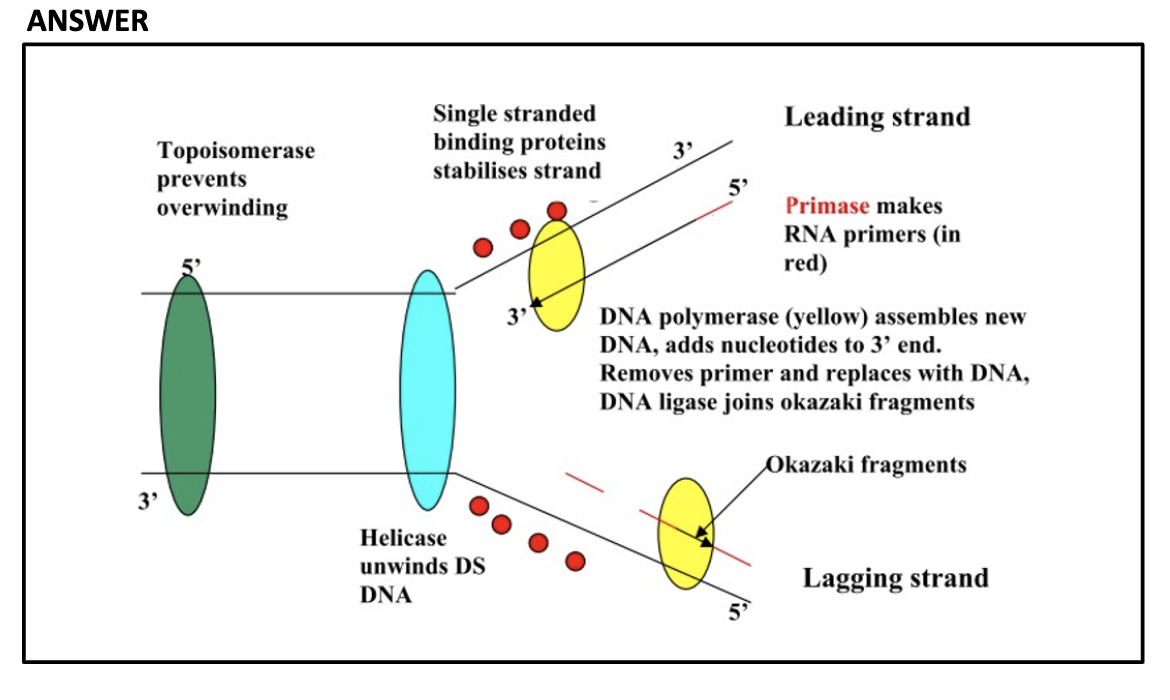
Explain the roles of DNA ligase, primers, primase, helicase, topoisomerase and single-strand binding proteins.
DNA ligase glue each okazaki fragments.
Primers indicate the location to DNA polymerase where to attach and synthesis nucleotide
Topiomerase unwind double helix structure prevent it from super coiling
Helicase breaks the H bonding between double helix DNA structure
SSBP binds to single strand of DNA prevent it from closing
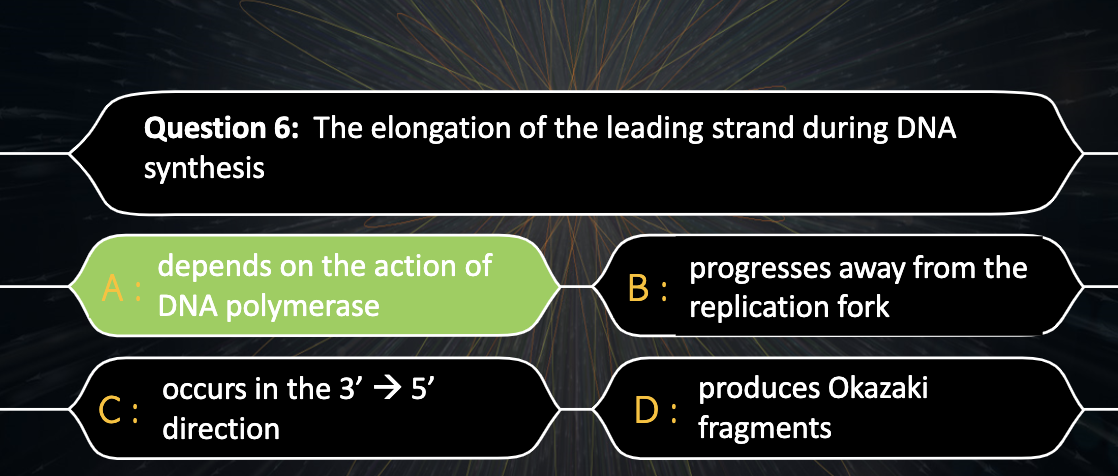
Describe the antiparallel nature of DNA and explain why continuous synthesis of both DNA strands is not possible.
DNA polymerase only attach nucleotides to OH group in 3' carbon, so the synthesis can only happen in 5'->3' direction. DNA is consist of two strands with antiparallel structure so only one of two strand can do continous synthesis
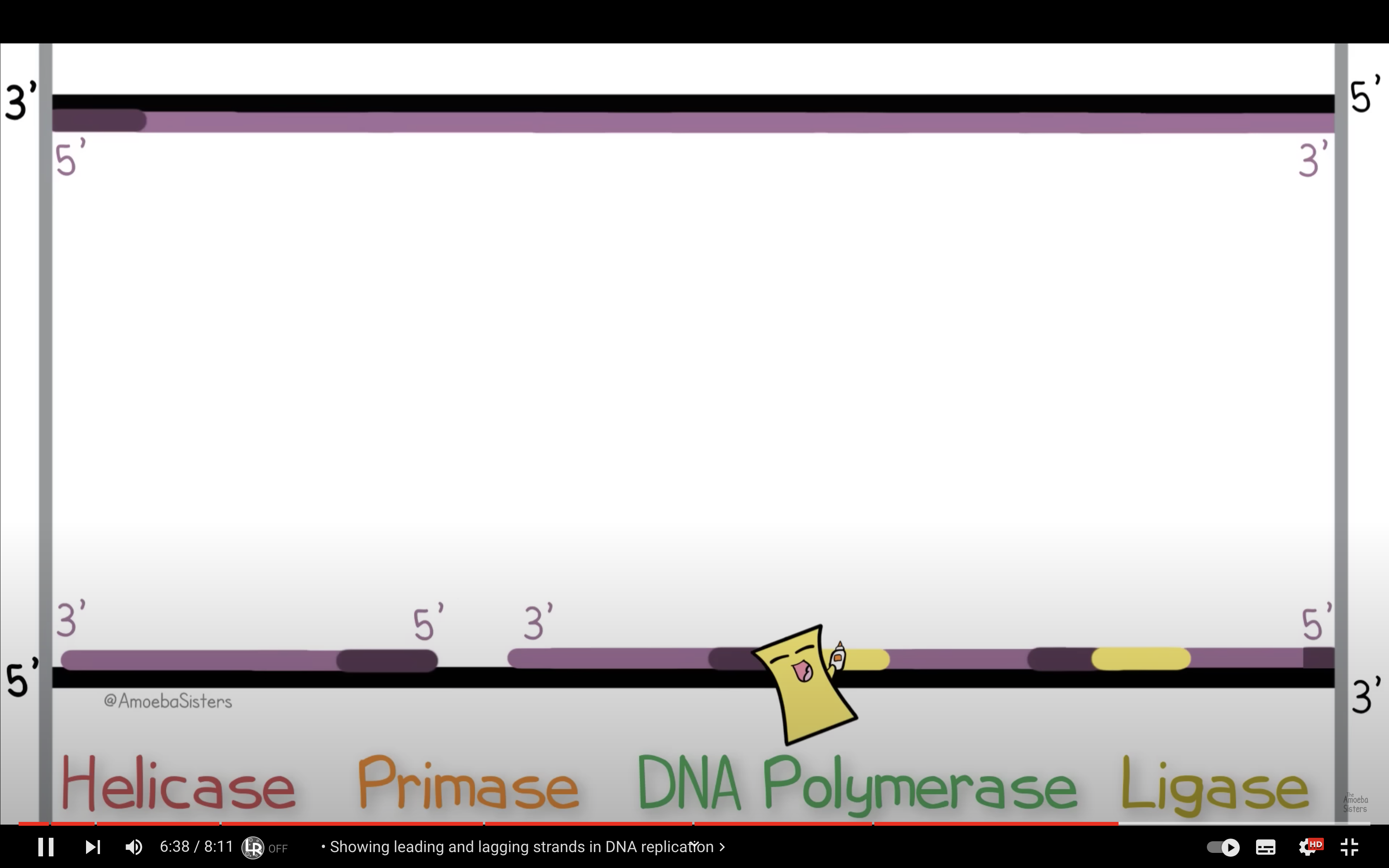
Distinguish between the leading strand and the lagging strand.
Leading strand is the one with 3'->5' direction , The replication fork and the DNA synthesis direction matches so the synthesis occurs continuously.
However, lagging strand has opposite direction with replication fork and DNA synthesis, making a DNA segments.
Explain how the lagging strand is synthesized even though DNA polymerase can add nucleotides only to the 3’ end.
DNA polymerase attach to the 3' end of the RNA primer and attach nucleotides to it, and when it arrives to the 5' end of the primer aheading it, then it dissassociate. Ligase glues the okazaki fragments together.
Describe the significance of Okazaki fragments.
They allow the lagging strand can syntheses While the leading strand synthesis DNA continuously.
Explain how mistakes are repaired in DNA replication and describe a disease/condition that arises as a result of errors in these pathways.
Mismatch Repair (MMR) - repair enzymes correct errors in base pairing, DNA can be damaged by exposure to harmful chemical or physical agents such as cigarette smoke and X-rays; can also undergo spontaneous changes
Nucleotide Excision Repair - a nuclease cuts out and replaces damaged stretches of DNA
If these errors aren't repaired, gene mutations can occur like cystic fibrosis.
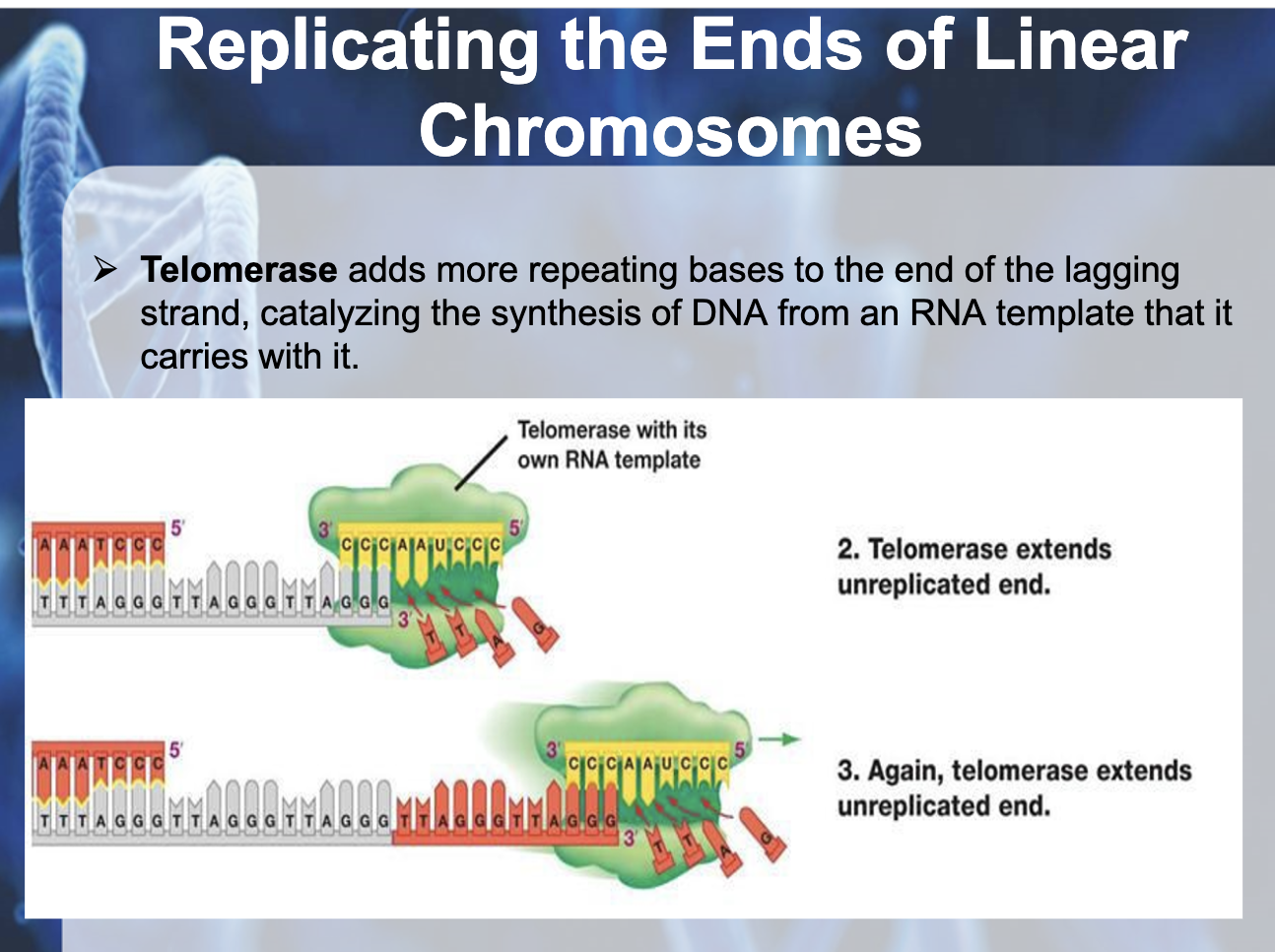
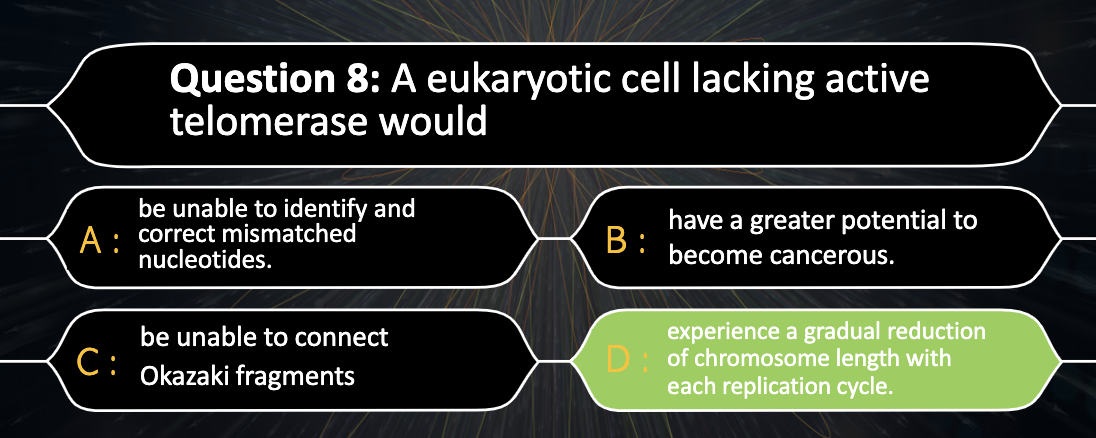
Understand why DNA replication in linear chromosomes can lead to shortening and how this is overcome in some cells.
Lagging strand has a short unreplicated end which degradesm shortening the chromosome after each replication.
In reproductive organs, telomerase extends the unreplicated end with its own RNA template. Somatic cells usually lack of telomerase and chromosome shortens by growing and aging. Telomerase activity in somatic cells has been linked to cancer
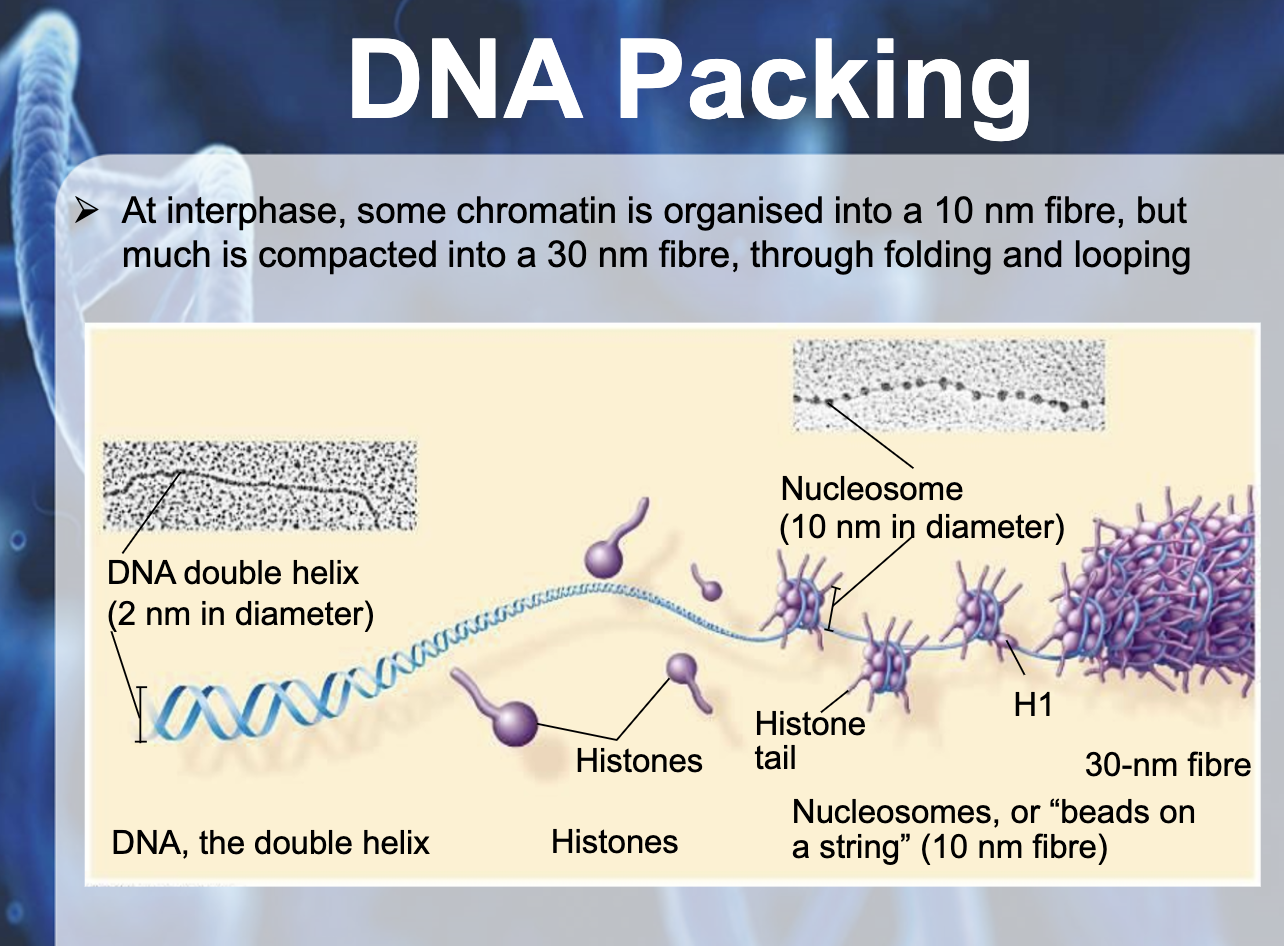
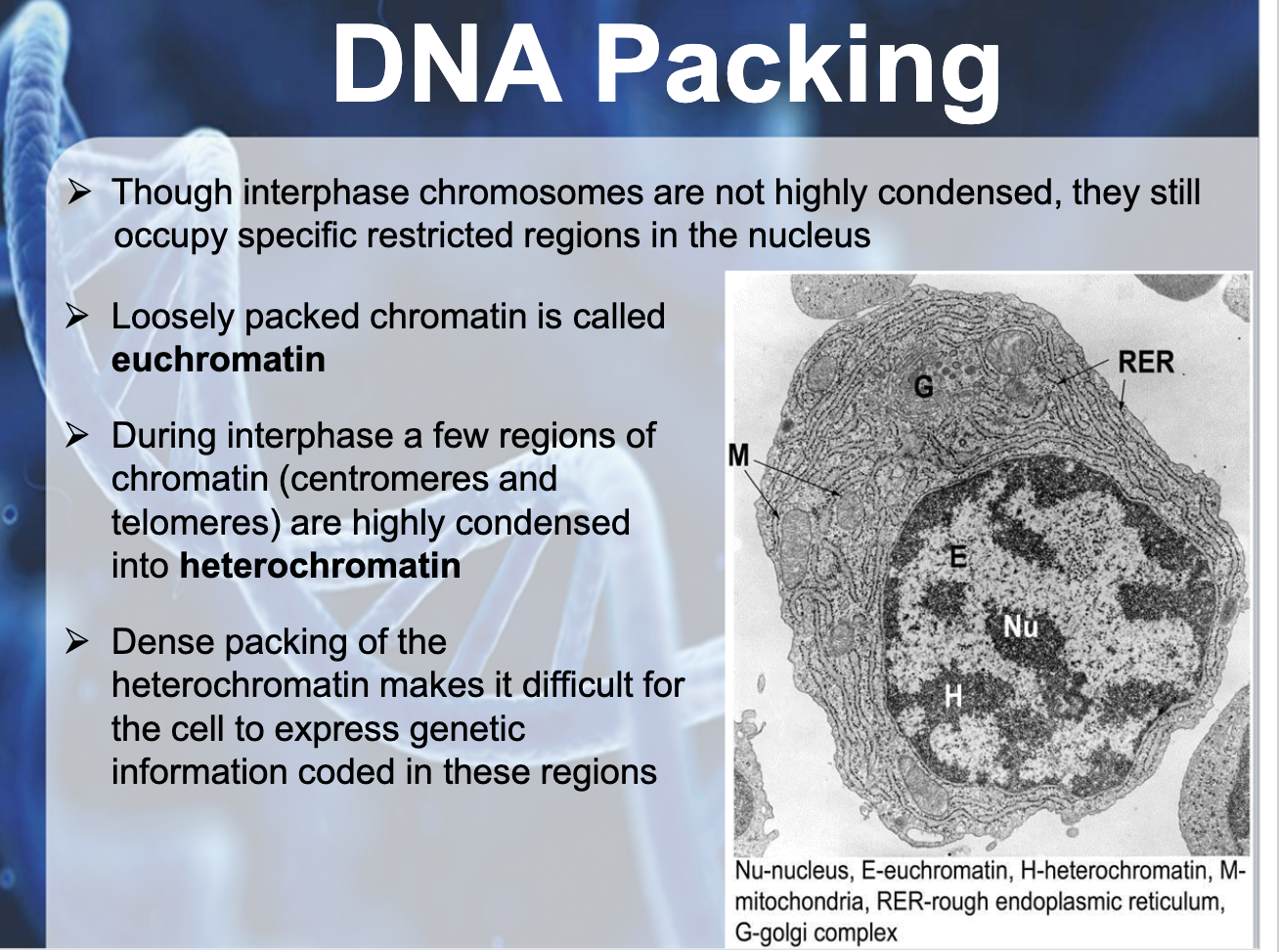
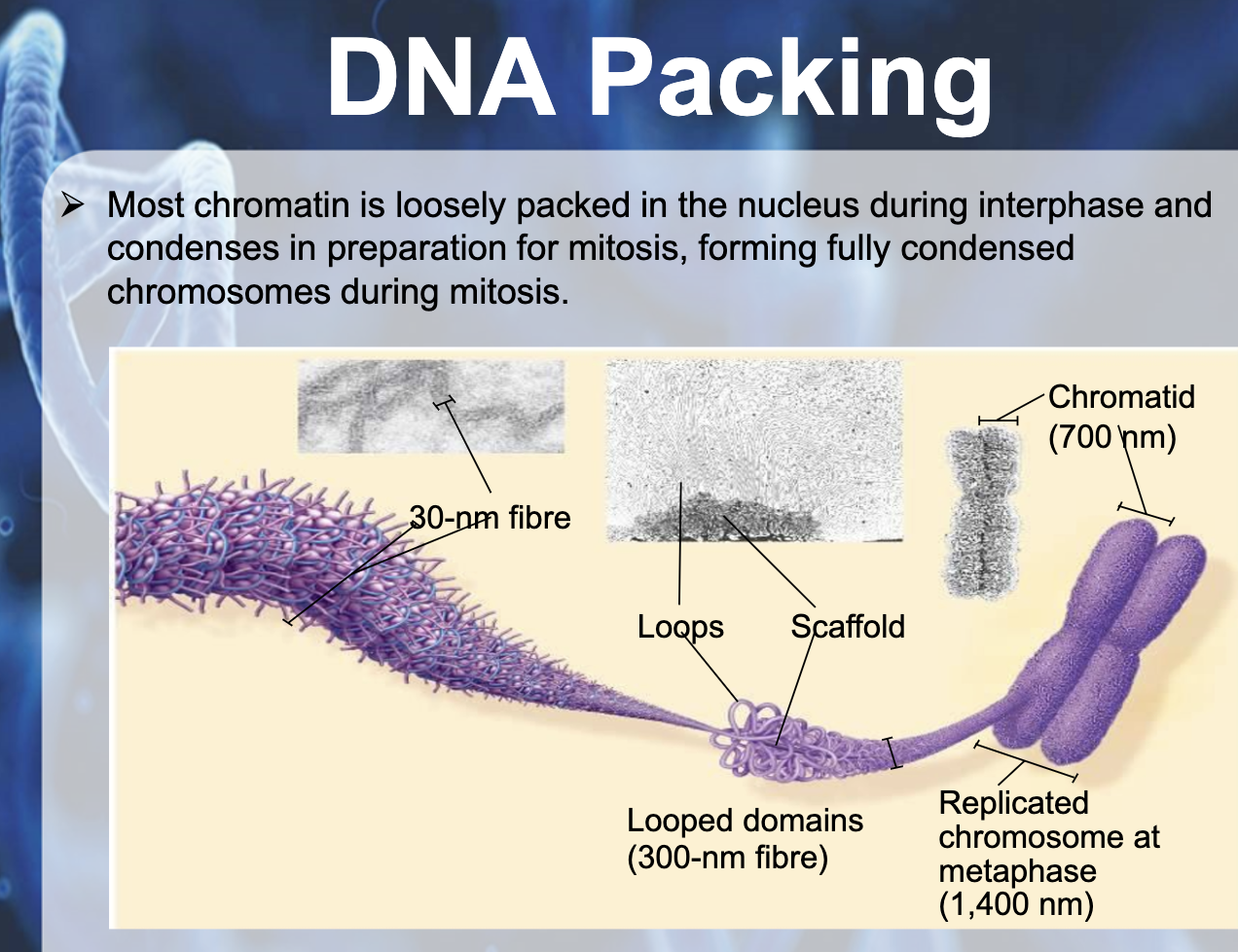
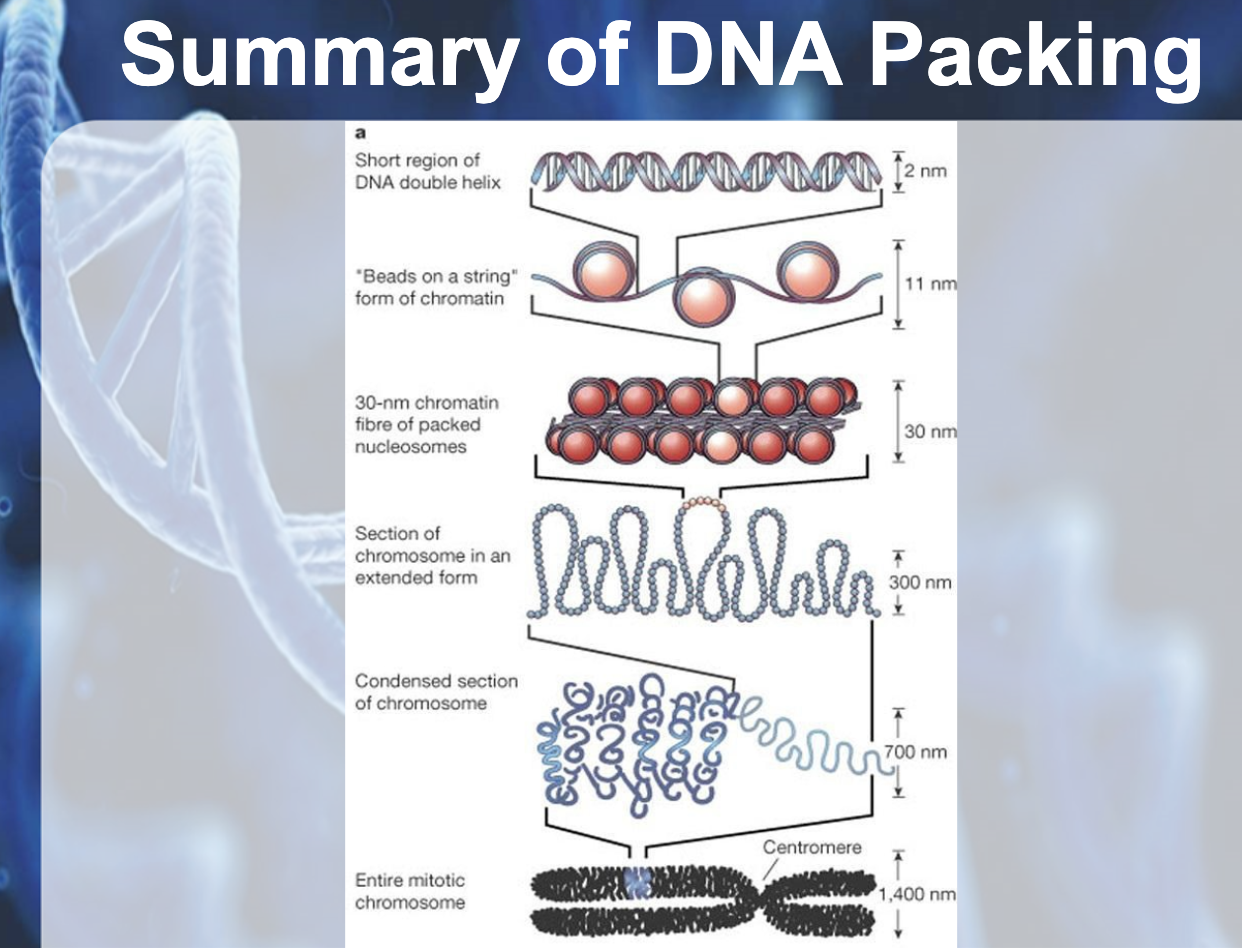
In bacteria (prokaryotic), DNA is supercoiled and found in nucleoid. In eukaryotic chromosomes, linear DNA molecules are associated with large amound of protein, histone. DNA is wrapped around the histone like a beads on a string. The 10nm fiber is called chromatin and the beads are termed nucleosome. Histone is positively charged and DNA is negatively charged (because of phosphate backbone), they can packaged together. They play a role in gene expression. Tightly packed area is called heterochromatin which doesn't contribute to gene expression, and the euchromatin is the area where gene expressed and it is untightly packed area.
PPT


THE SEARCH FOR THE GENETIC MATERIAL

-1869 Friedrich Miescher isolated nucleic acid from WBC nuclei

-Frederick Griffith began with research of the genetic role of DNA


DNA STRUCTURE
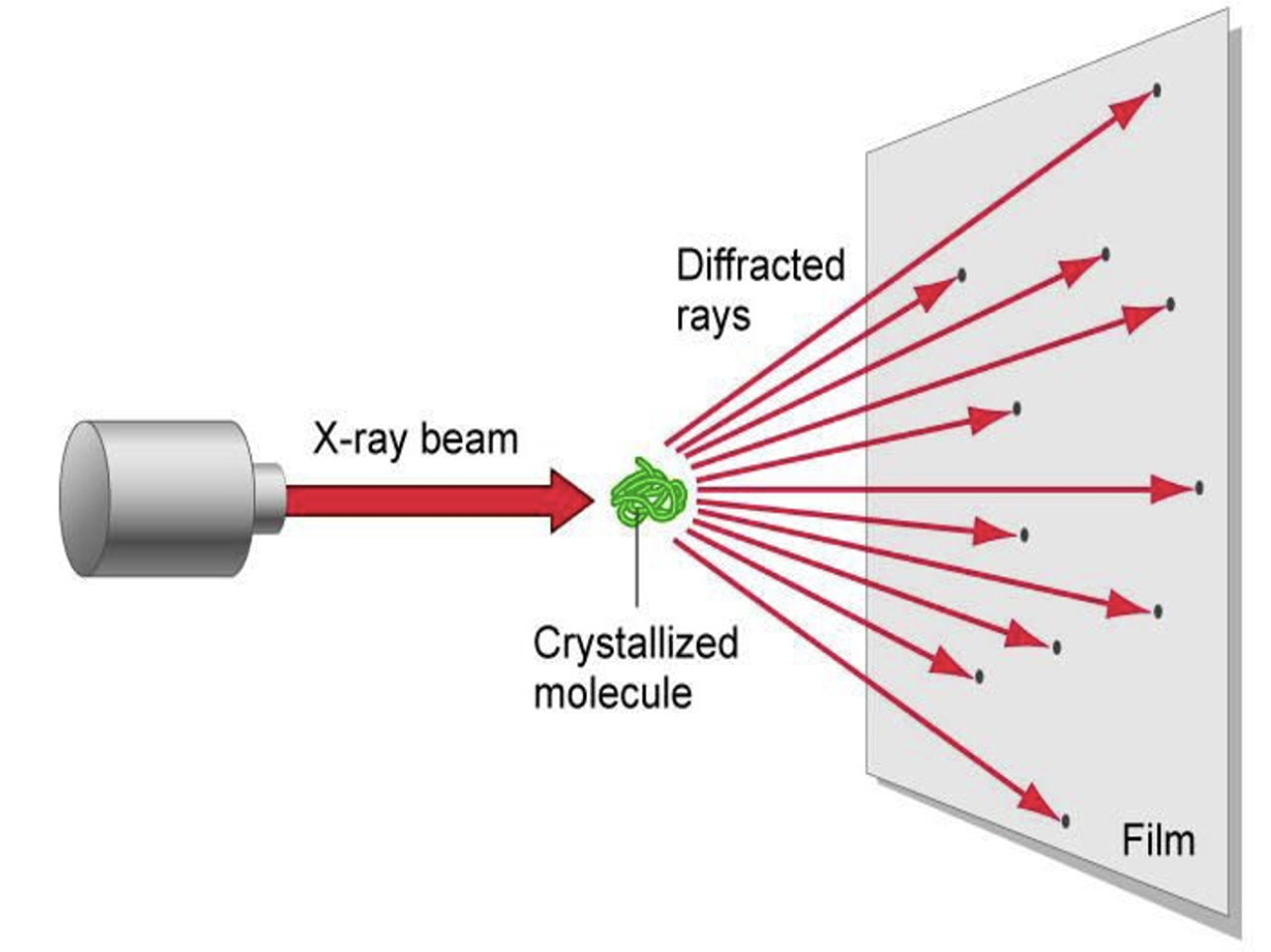
-James Watson & Francis Crick => Structure of DNA
-Maurice Wilkins & Rosalind Franklin => used X-ray crystallography to study molecular structure
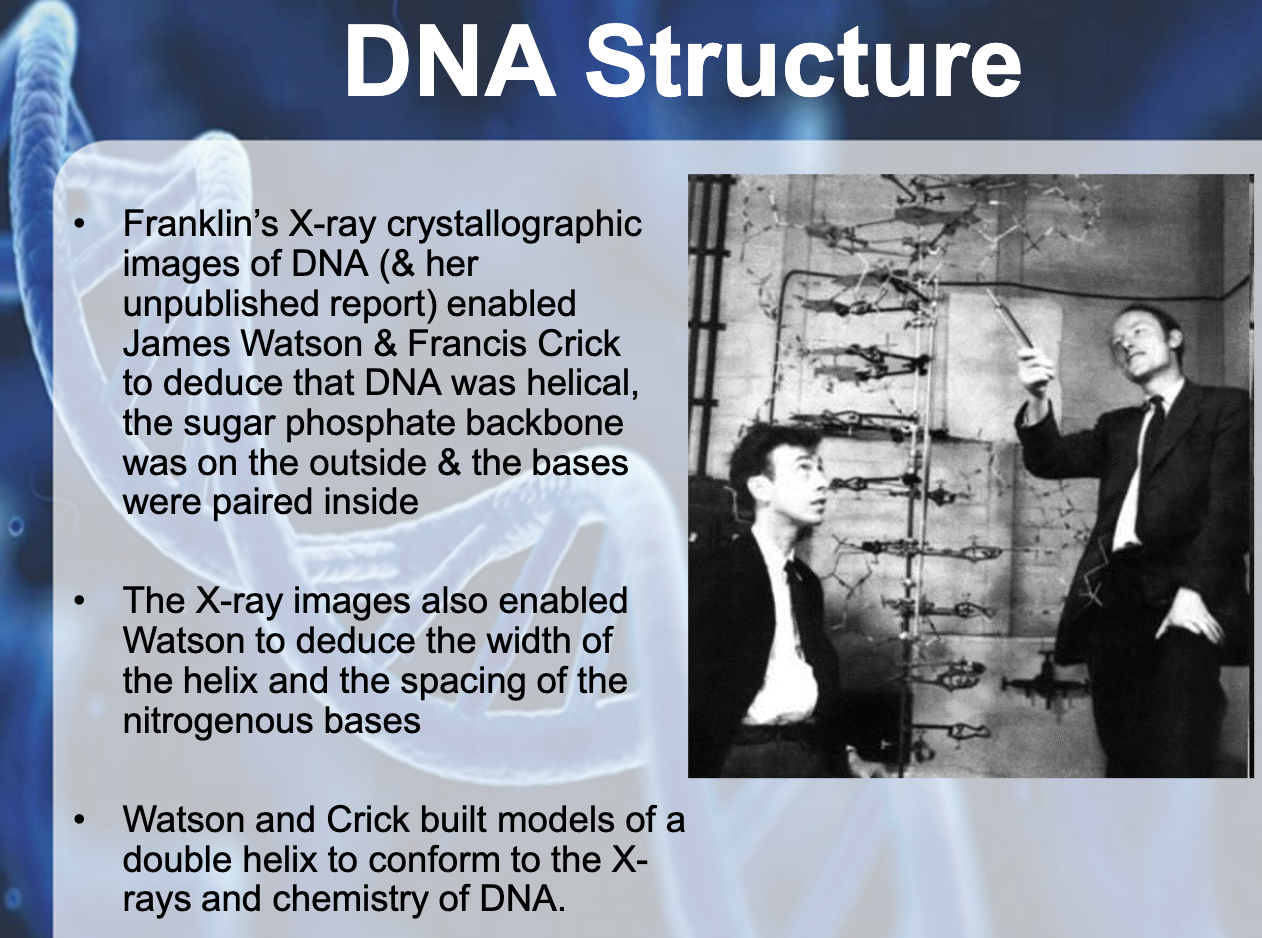
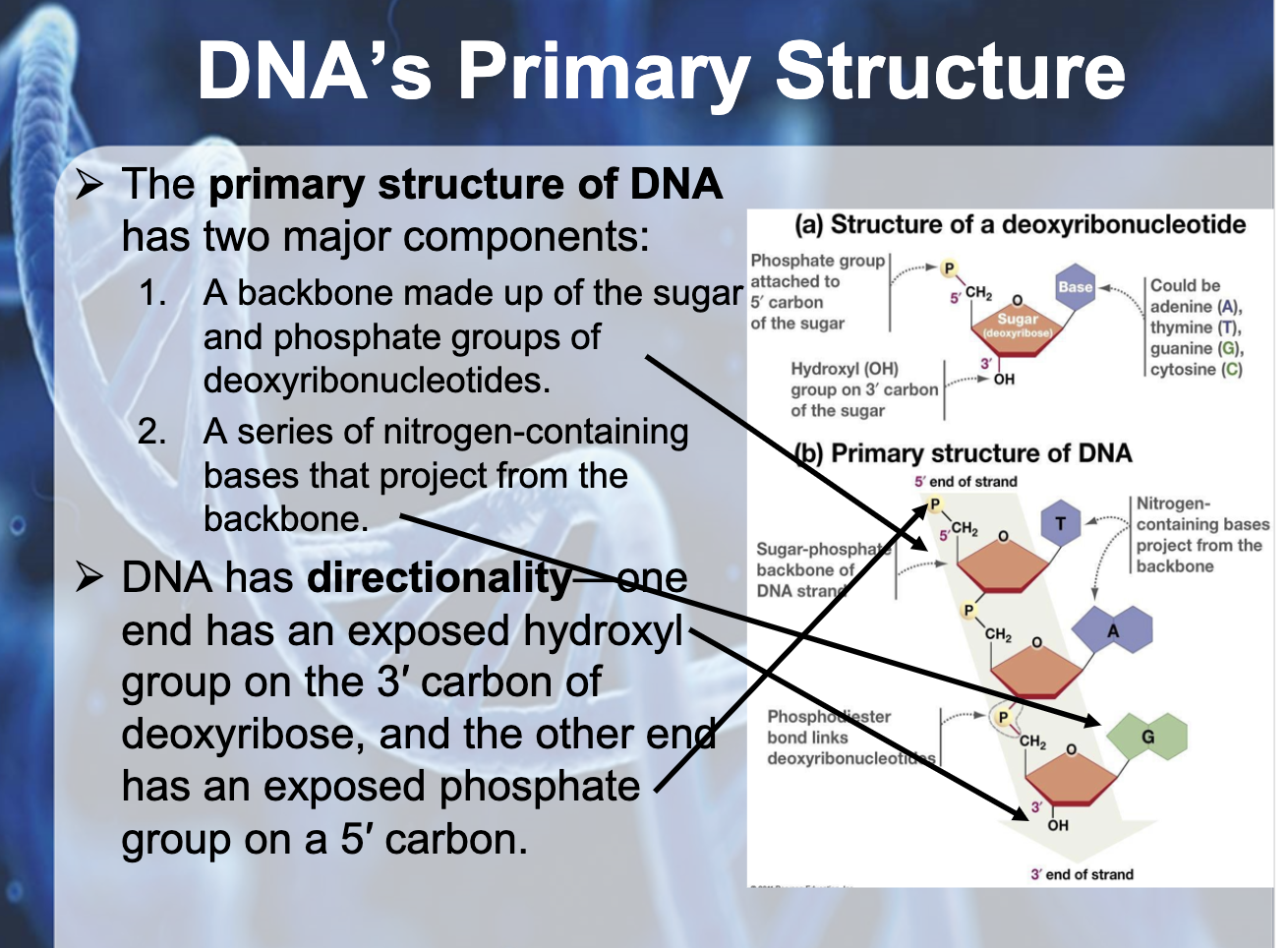
-Franklin's X-ray crystallographic images of DNA enabled James Watson & Francis Crick to deduce that DNA was helical (sugar phosphate backbone, sugar paired inside)
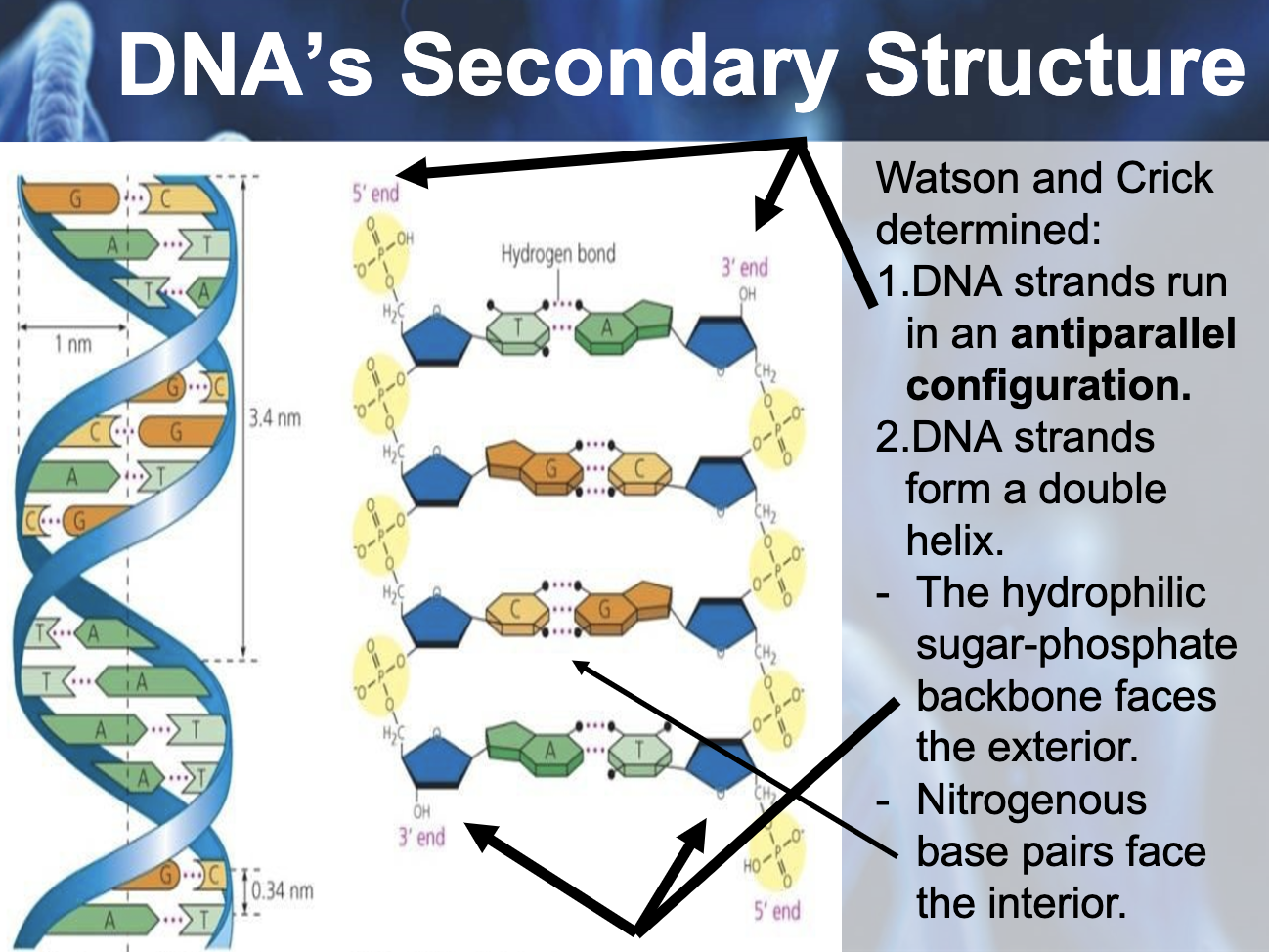
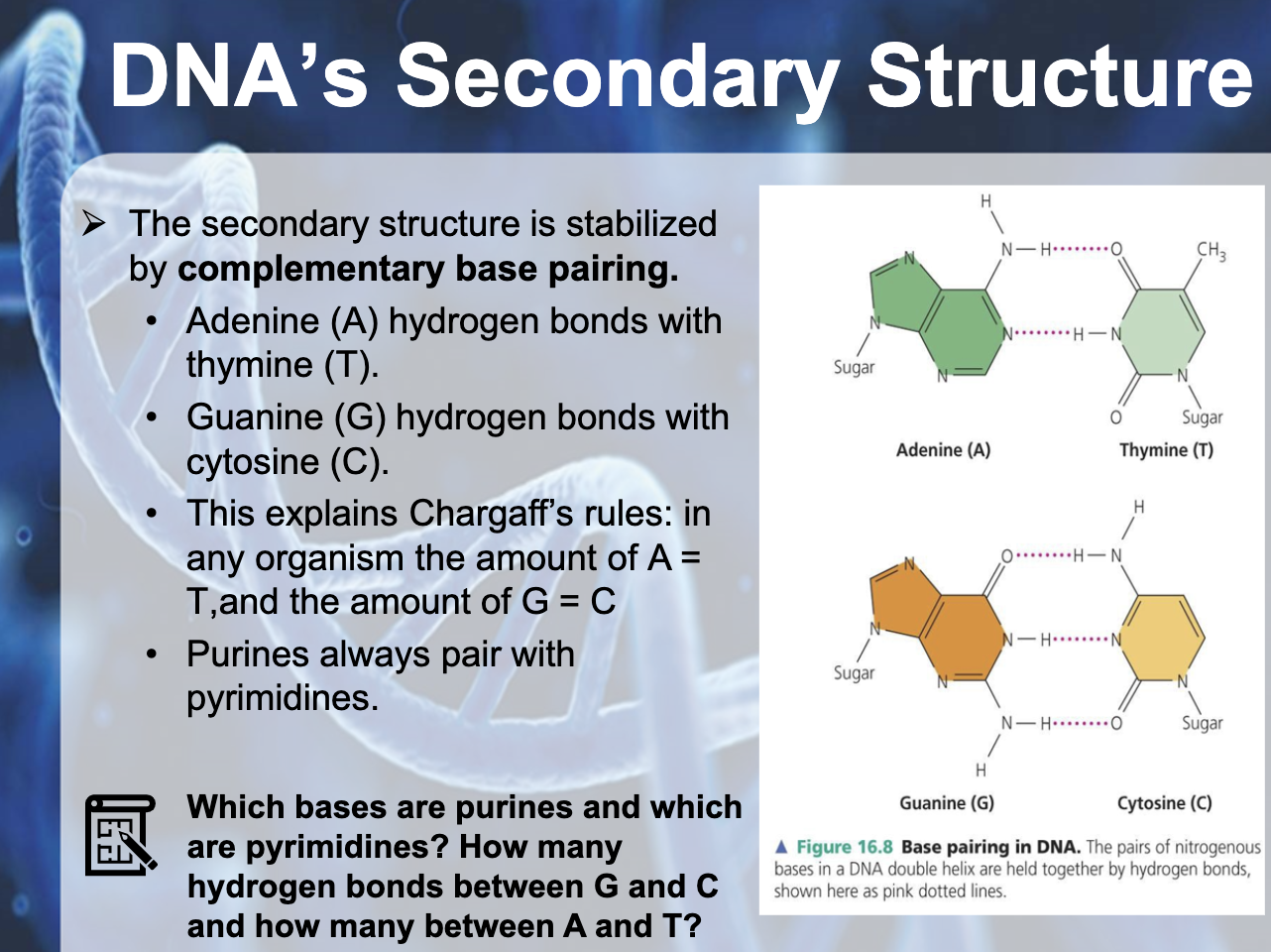
-Between A, T => 2 hydrogen bonding
-Between G, C => 3 hydrogen bonding
https://www.youtube.com/watch?v=o_-6JXLYS-k
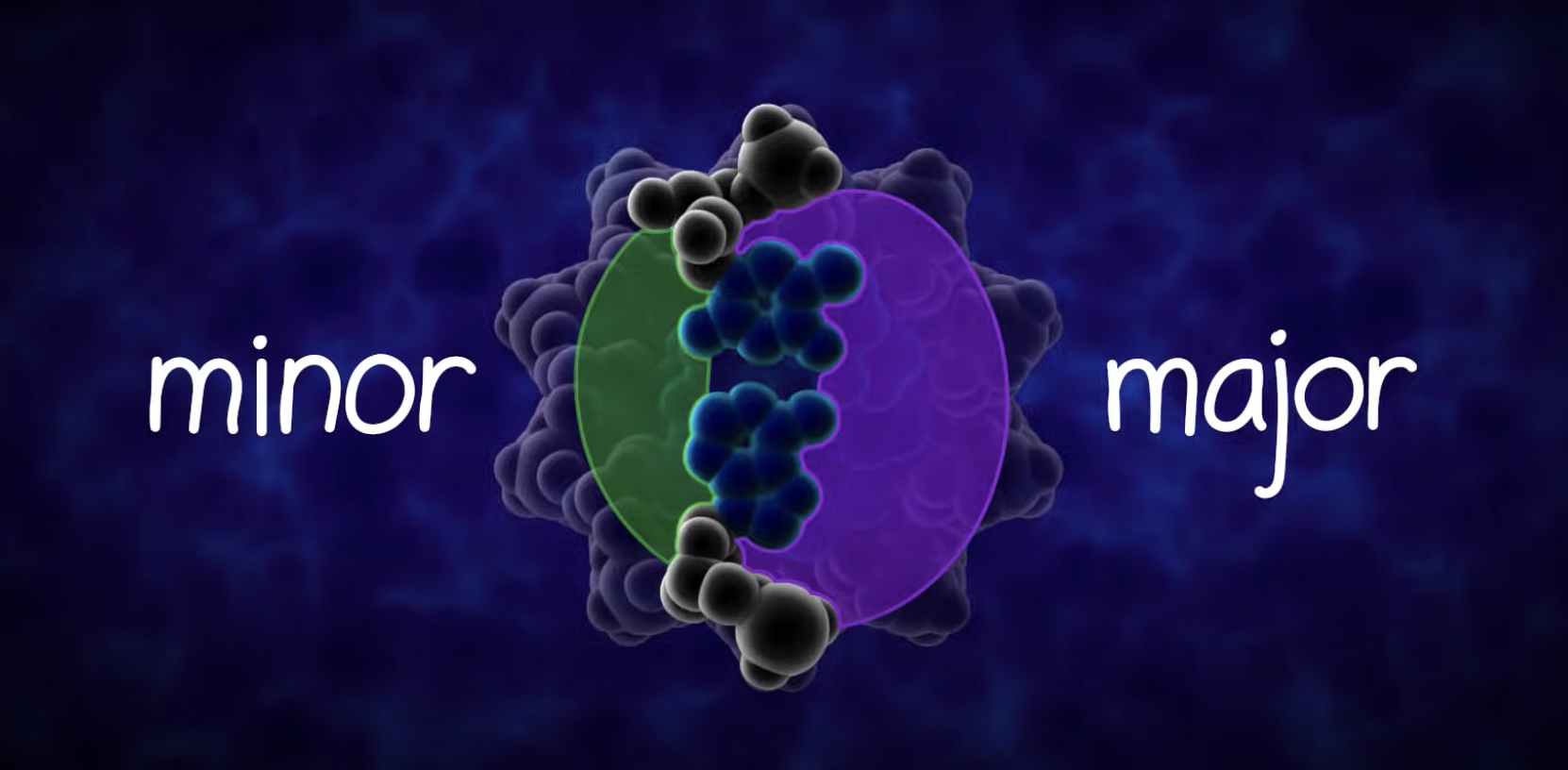
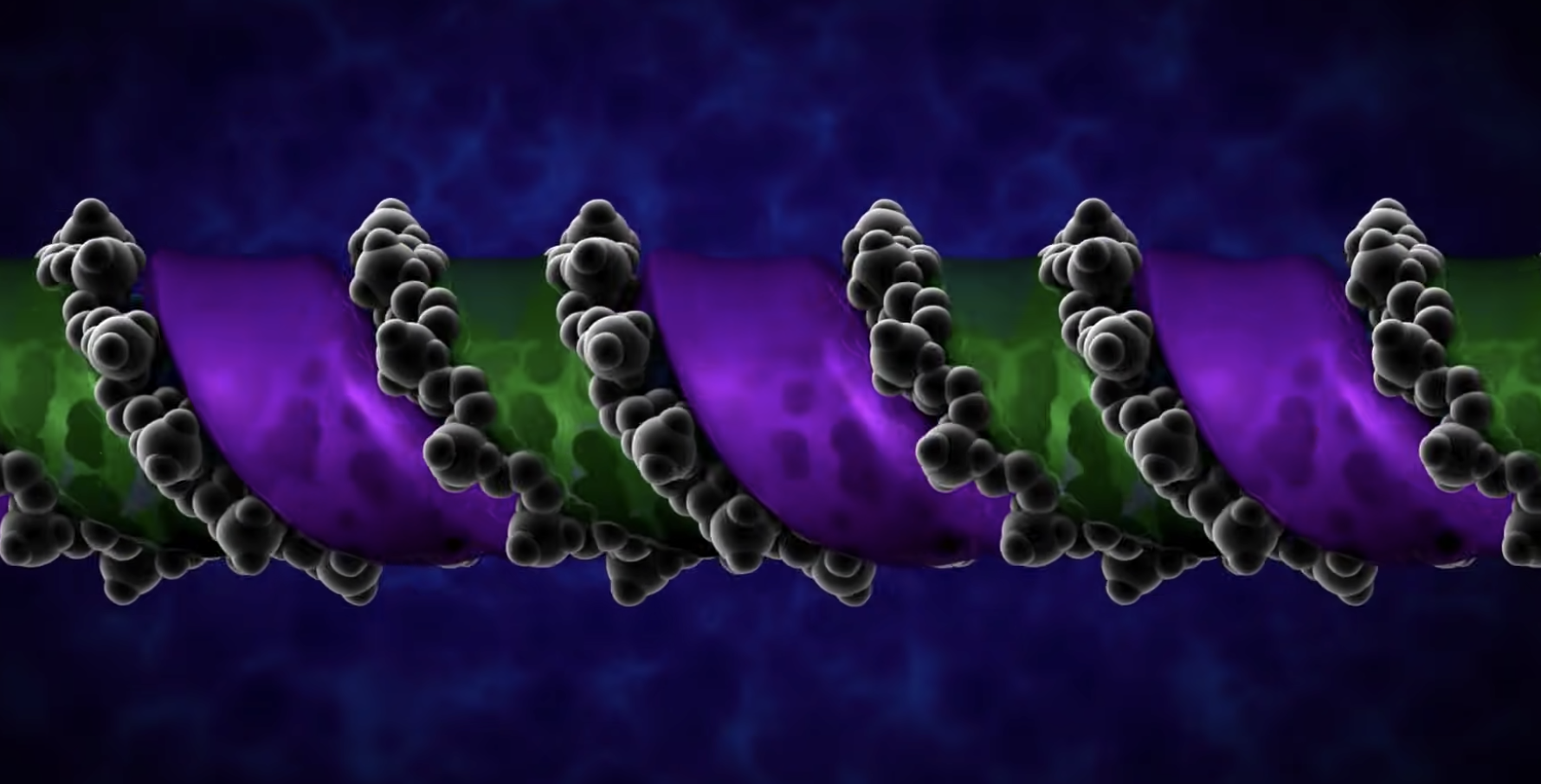
DNA REPLICATION

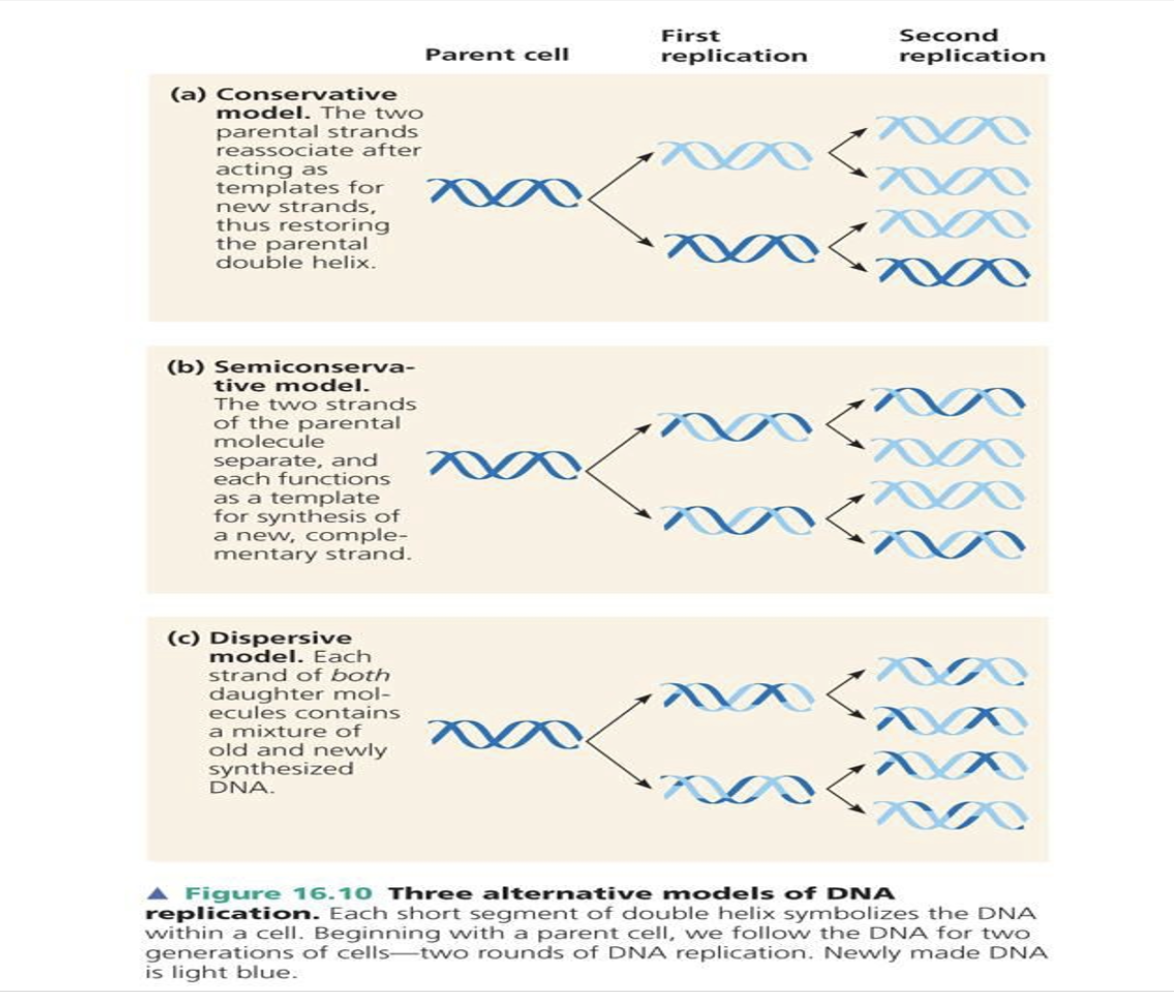
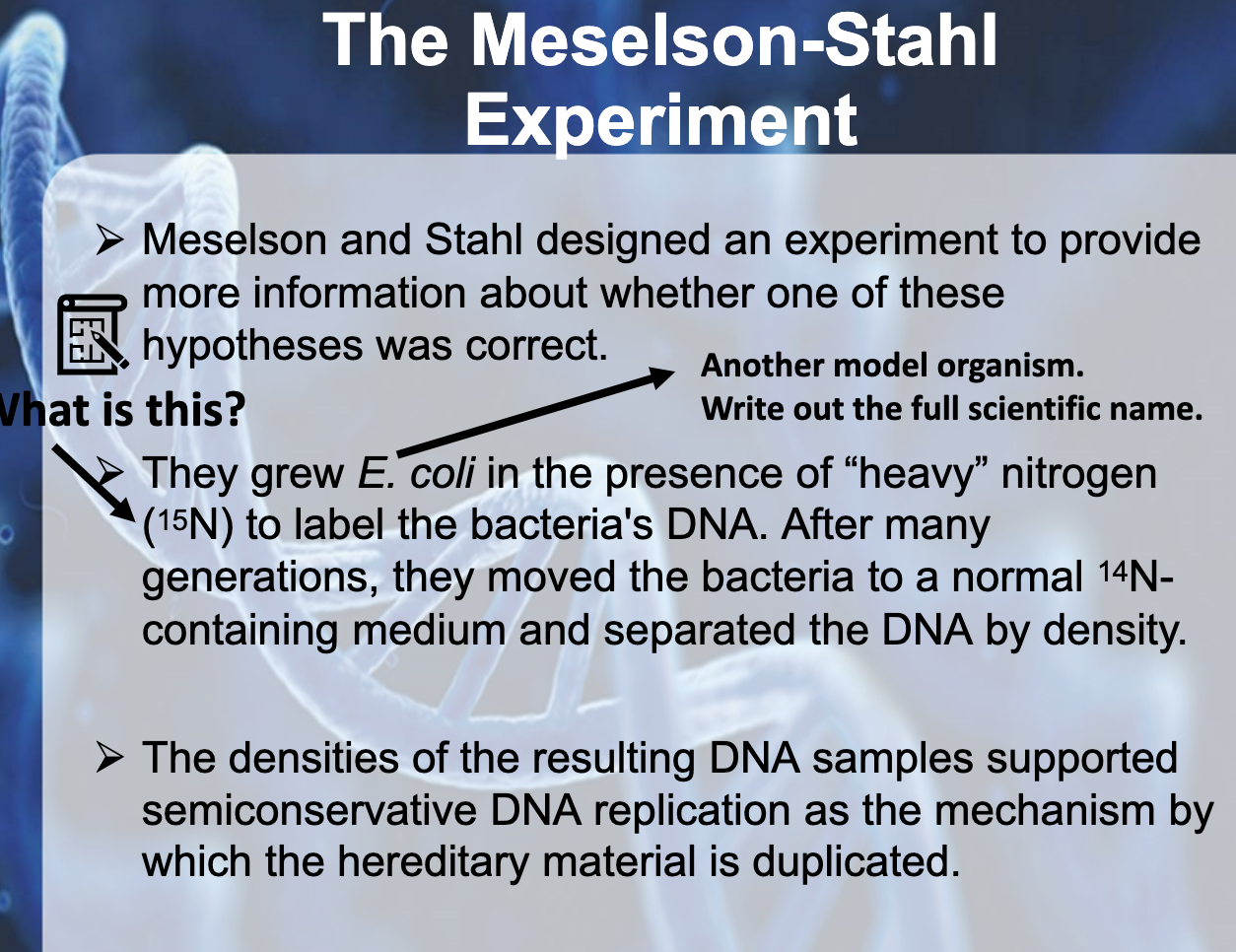
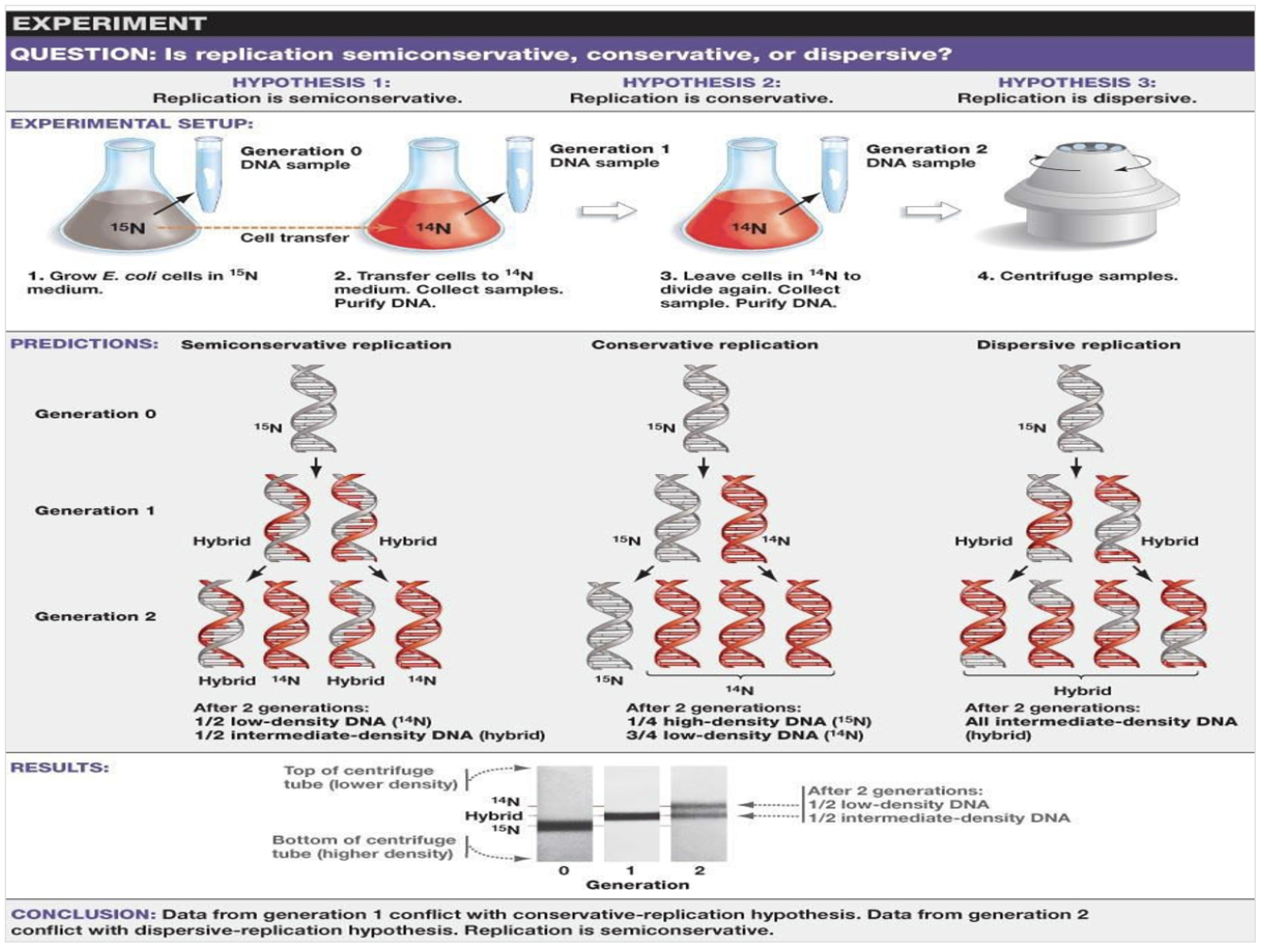
When they start with N15 DNA strands, They could only observe hybrids of DNA strands in G1. If it was semi- conservative model, they should observe N15 and N14 of DNA strands. In G2, They could observe N14 and hybrid in 1:1 ratio. If it was hybrid model, all of the DNA strands should hybrid strands.
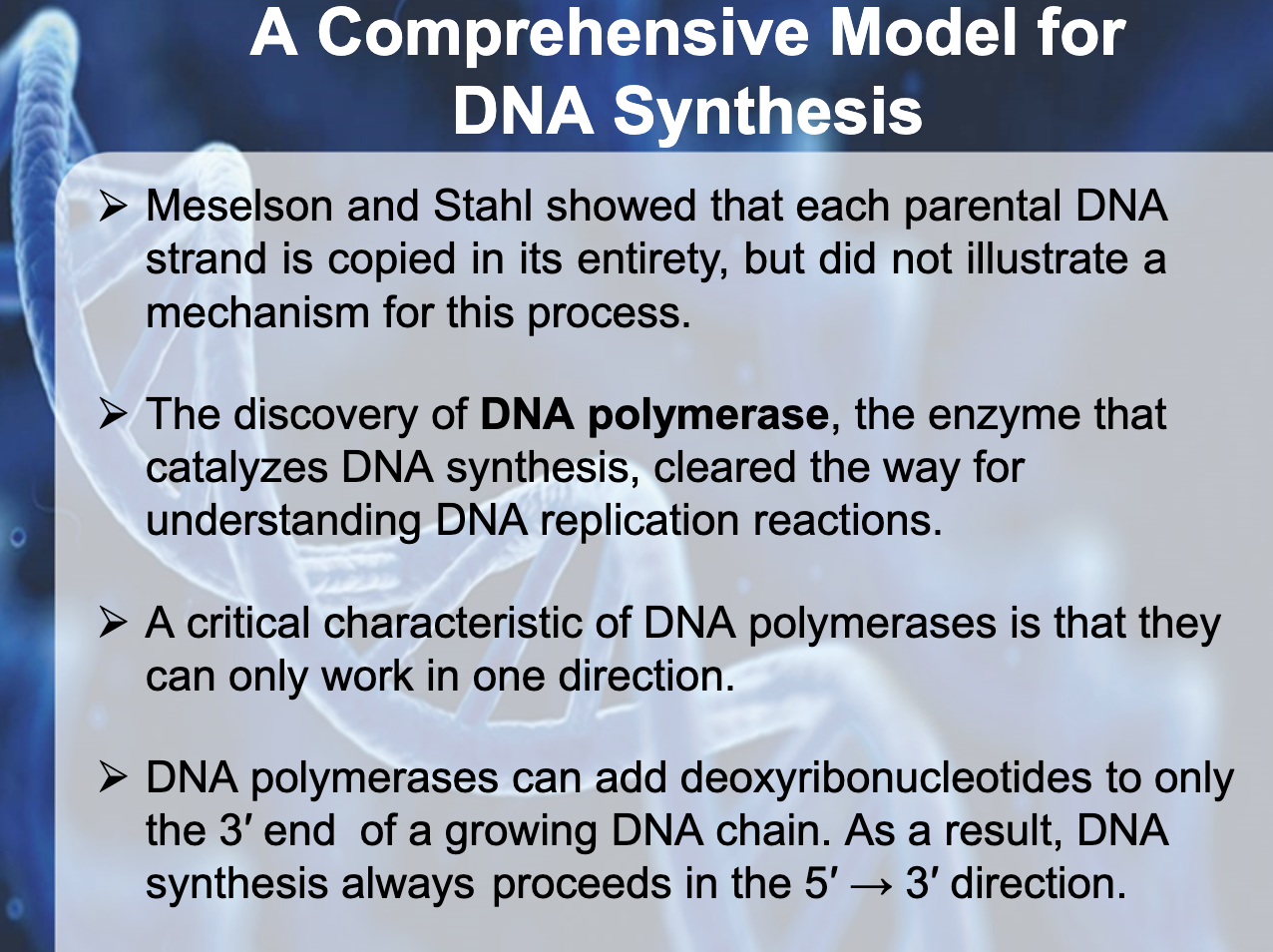
-DNA polymerase : enzyme that catalyzes DNA synthesis
(They can only work in one direction/ DNA synthesis always proceeds in the 5'->3' direction)
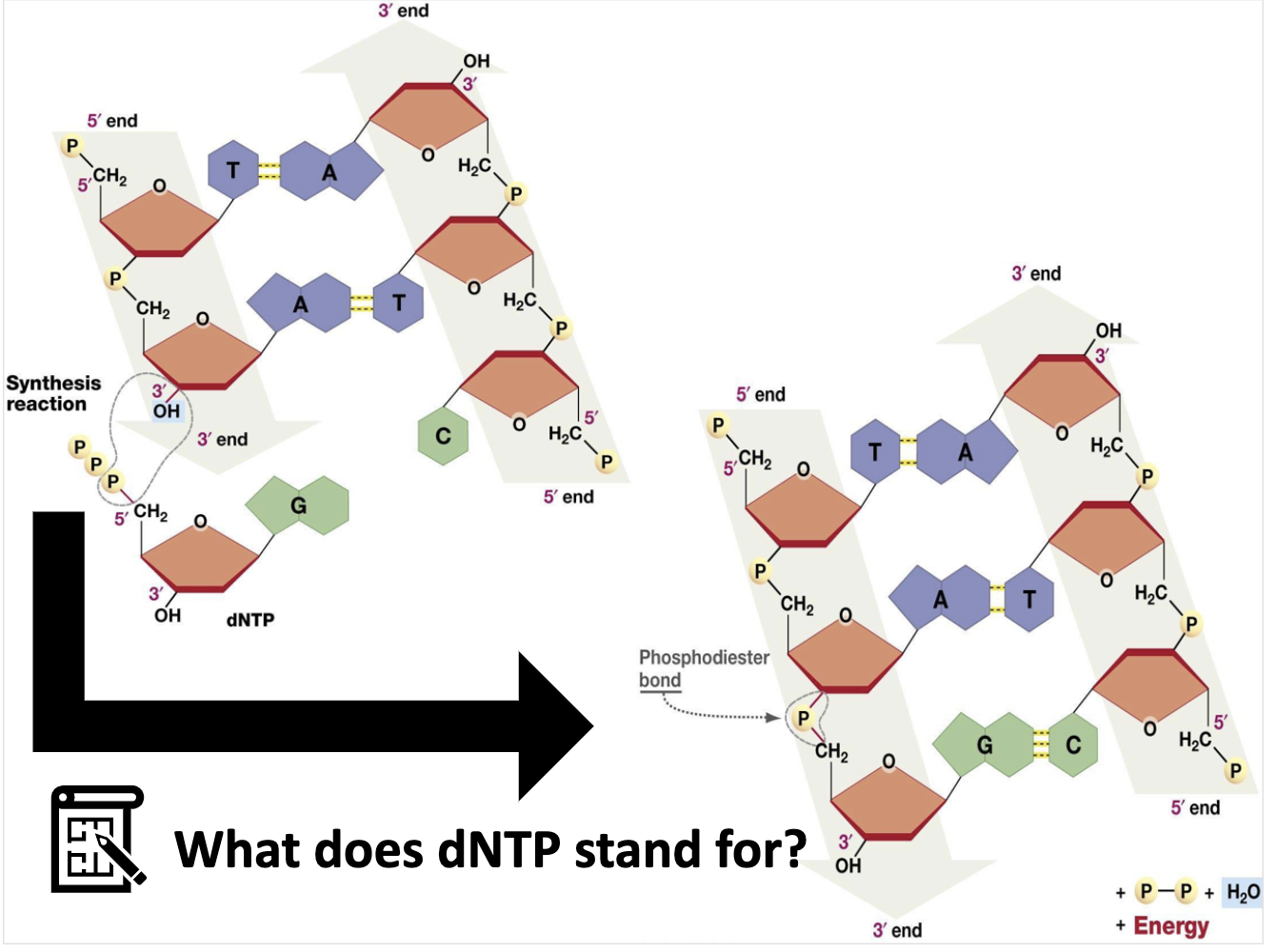
*dNTP => deoxynucleoside triphosphate
(nucleotide with deoxyribose)
*NTP => nuceloside triphosphate
*Nuceloside = ribose + base
*Nucleotide = ribose + base + triphosphate ( nuceloside + triphosphate )
*NTP => RNA / dNTP => DNA
(NTP -> ATP,TTP,GTP,CTP / dNTP -> dATP, dTTP, dGTP, dCTP)
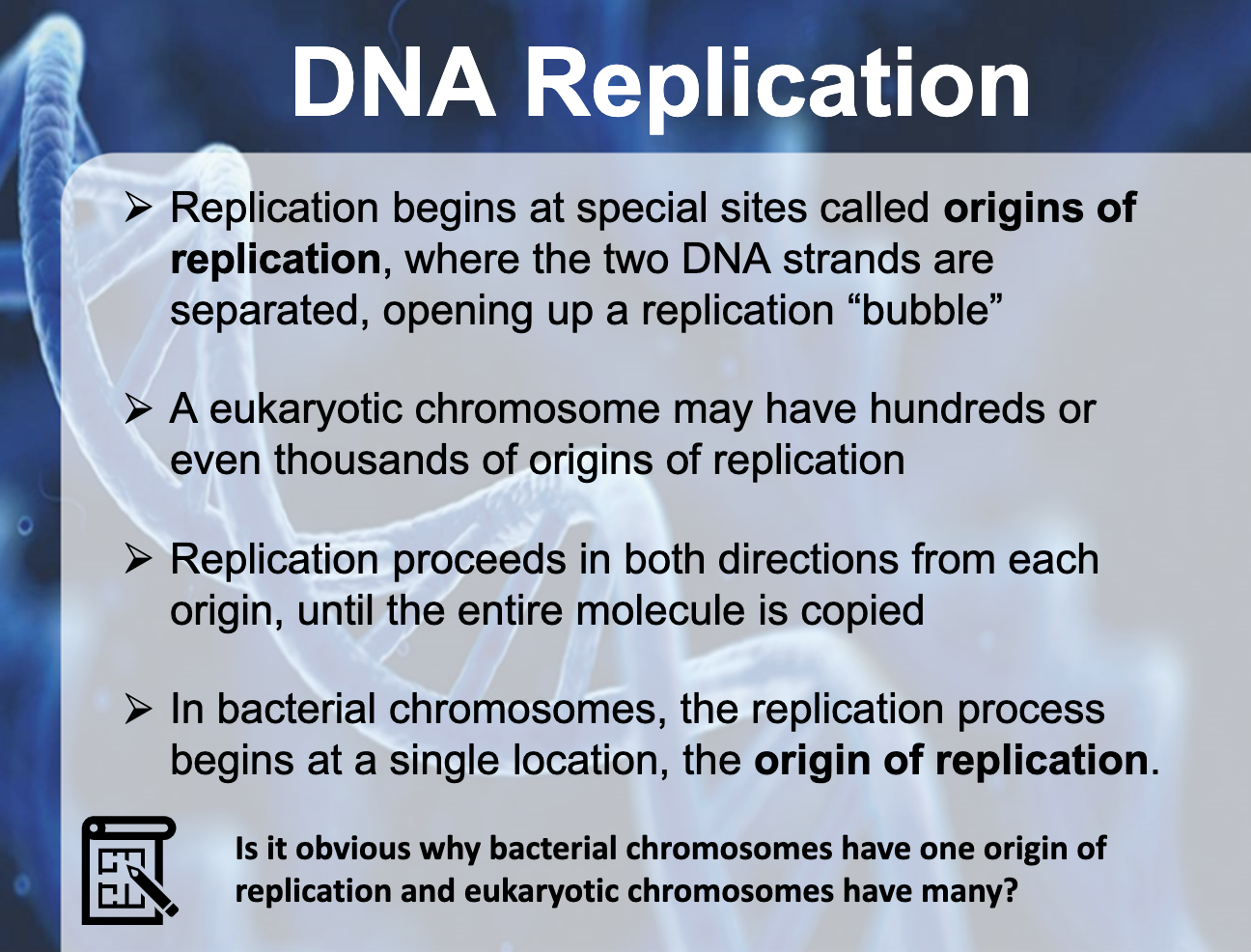
*Why bacterial chromosomes have one origin of replication and eukaryotic chromosomes have many?
Eukaryotes typically have multiple linear chromosomes, each with many origins. Multiple origins are a necessity for eukaryotes as they have much larger genomes than bacteria and eukaryotic replication forks move about 20 times more slowly than bacterial replication forks.
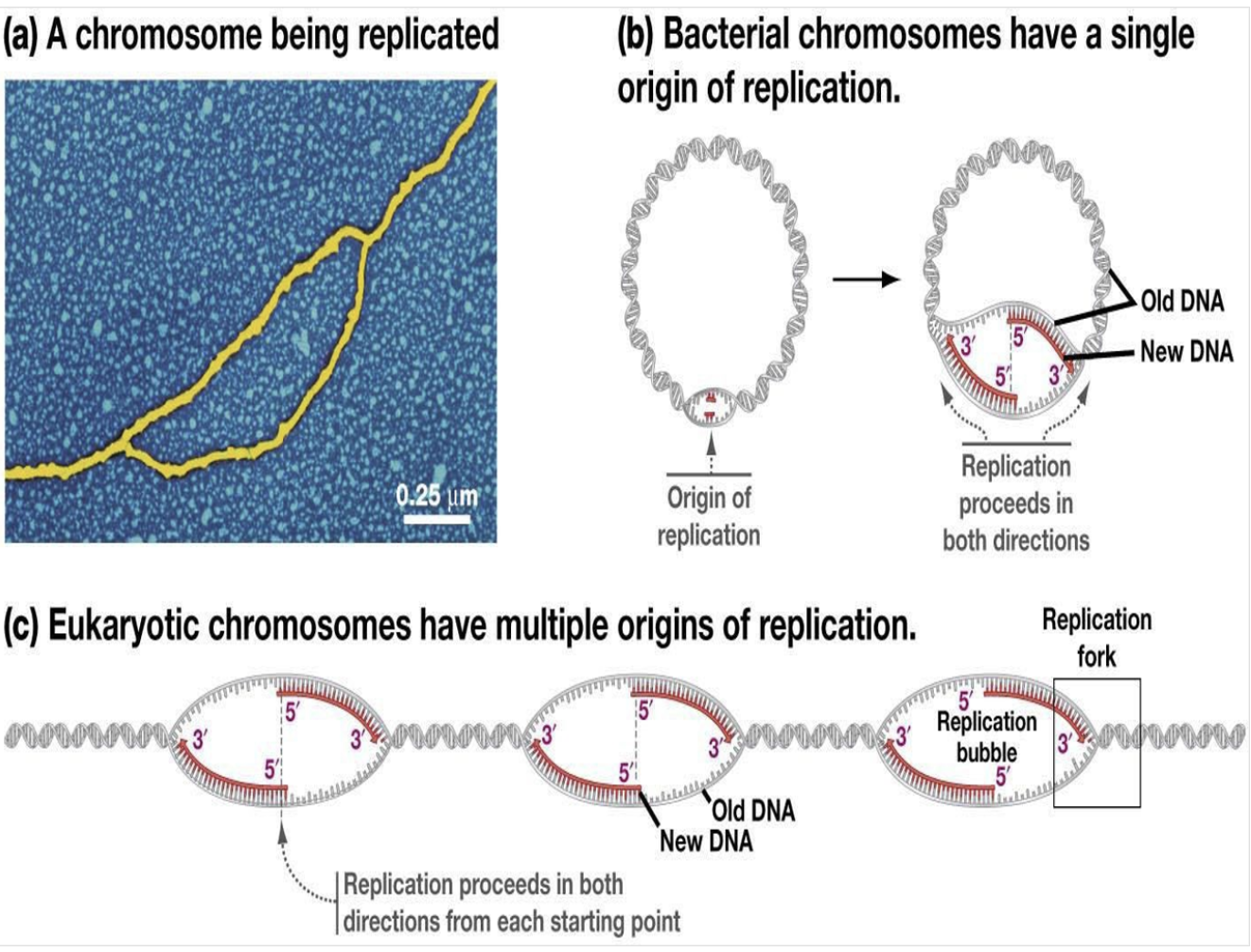
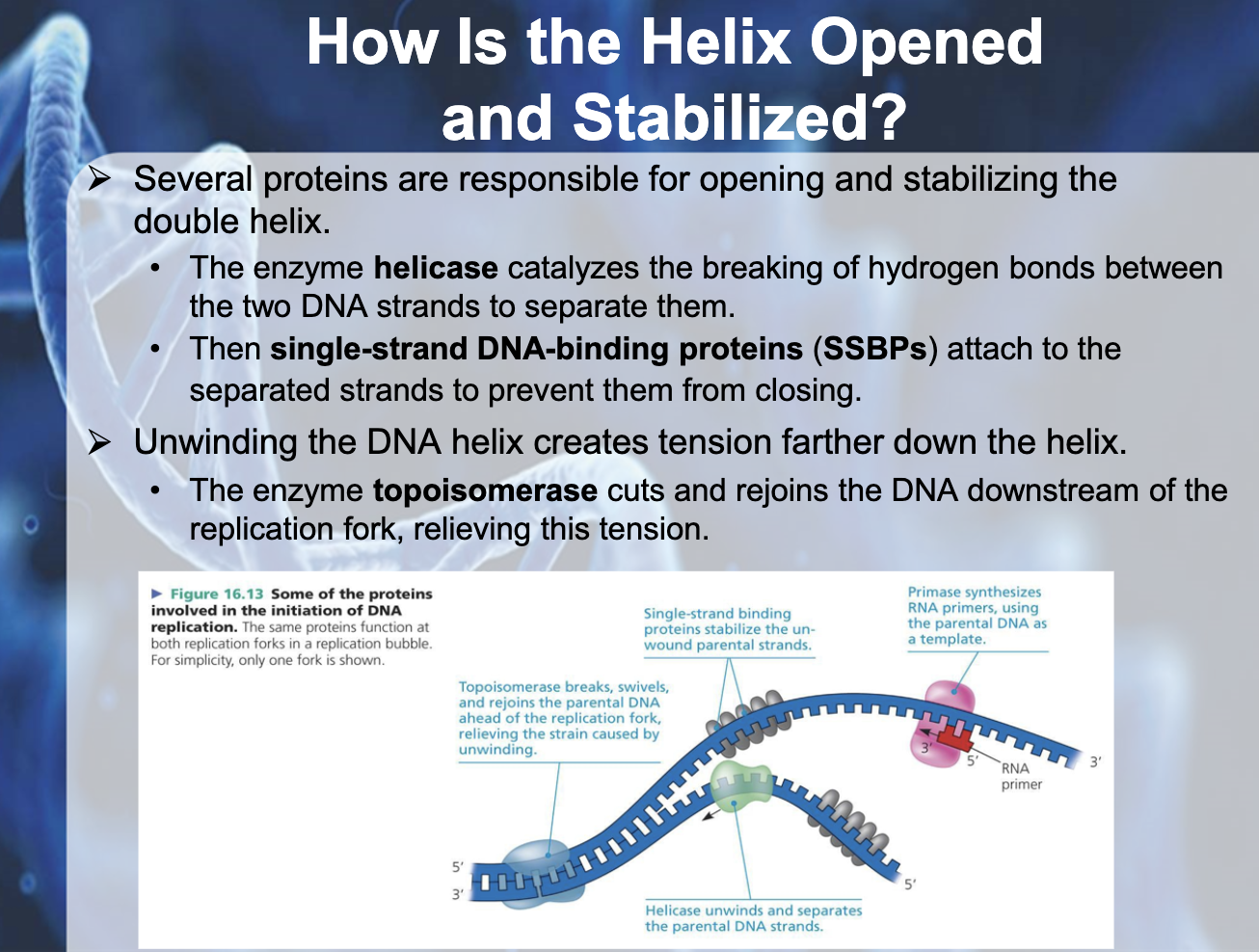
*Helicase breaks the hydrogen bonding between the two DNA strands to separate them
*Single-strand DNA binding proteins (SSBPs) attach to the separated strands to prevent them from closing
*Topoisomerase cuts and rejoins the DNA downstream of the replication fork, relieving tension
*Topoisomerase unwind the double helix structure, Helicase breaks the H bonding between the DNA strands, SSBP binds to each strand of DNA to prevent it from binding again.(prevent it from closing)
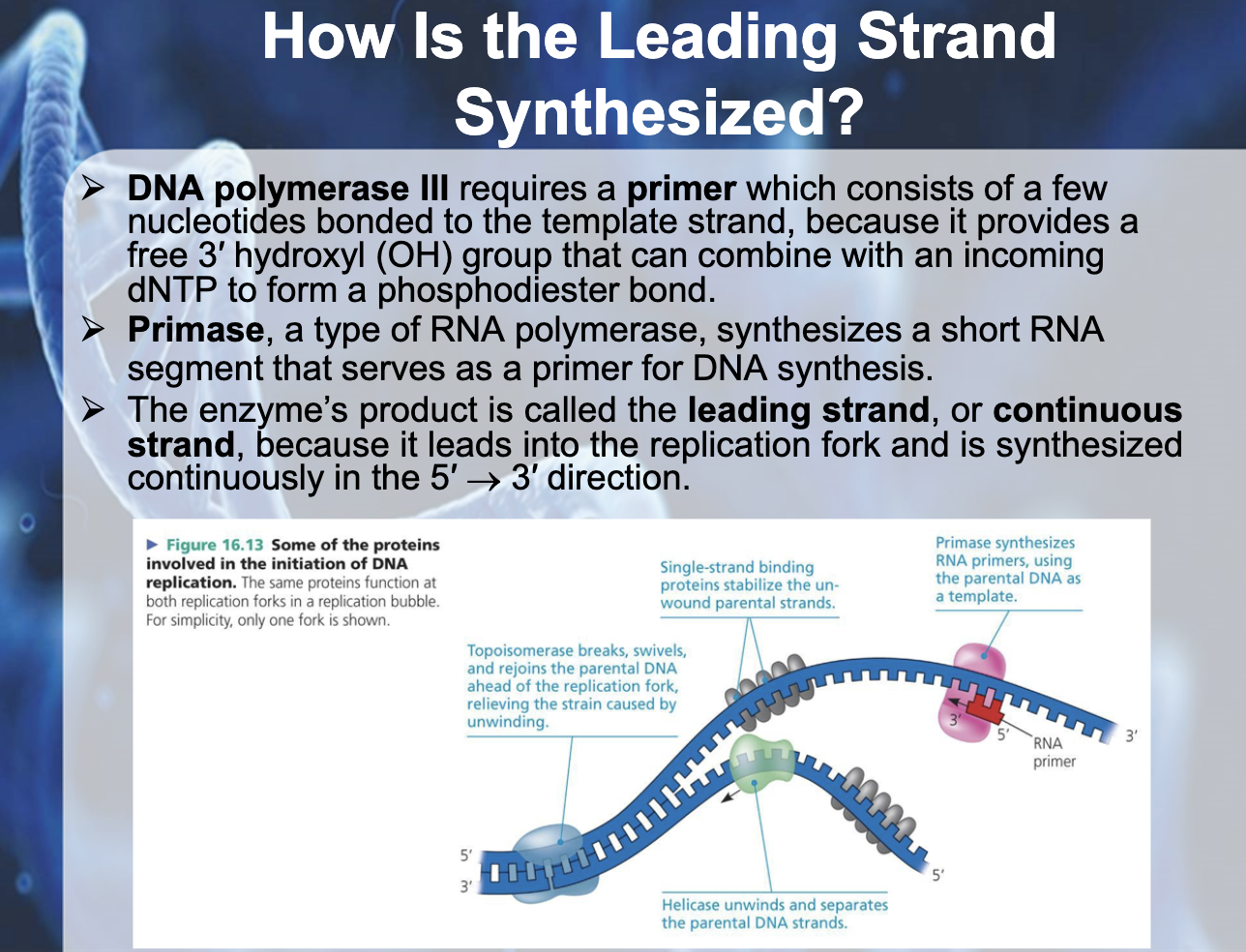
프라이머(primer) 또는 시발체(始發體)는 DNA 합성(중합)의 기시점이 되는 짧은 유전자 서열
-DNA polymerase III : replicate the genome in order to ensure the maintenance of the genetic information and its faithful transmission through generations.
*DNA polymerase3 combine dNTP to the C3' OH group forming phosphodiester bond
*Primase binds to the single strand DNA and synthesis RNA primer, DNA polymerase attach to the 3' end of the primer and syntheses DNA fragment. When it arrives to the 5' end of other primer then it dissassociate. Ligase glue okazaki fragments together
-Primer : consists of a few nucleotides bonded to the template strand,
-Primase : type of RNA polymerase, synthesizes a short RNA segment that serves as a primer for DNA synthesis
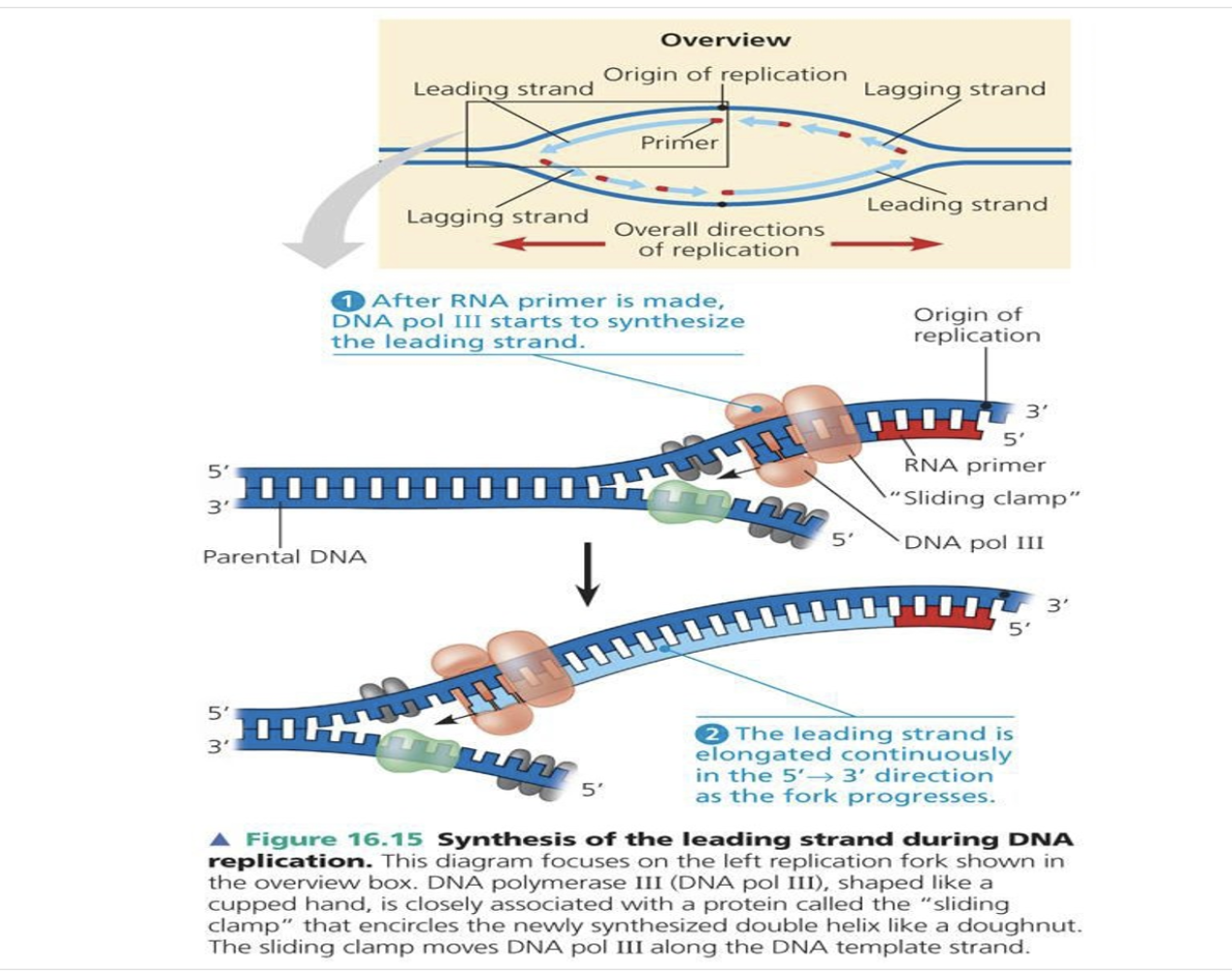
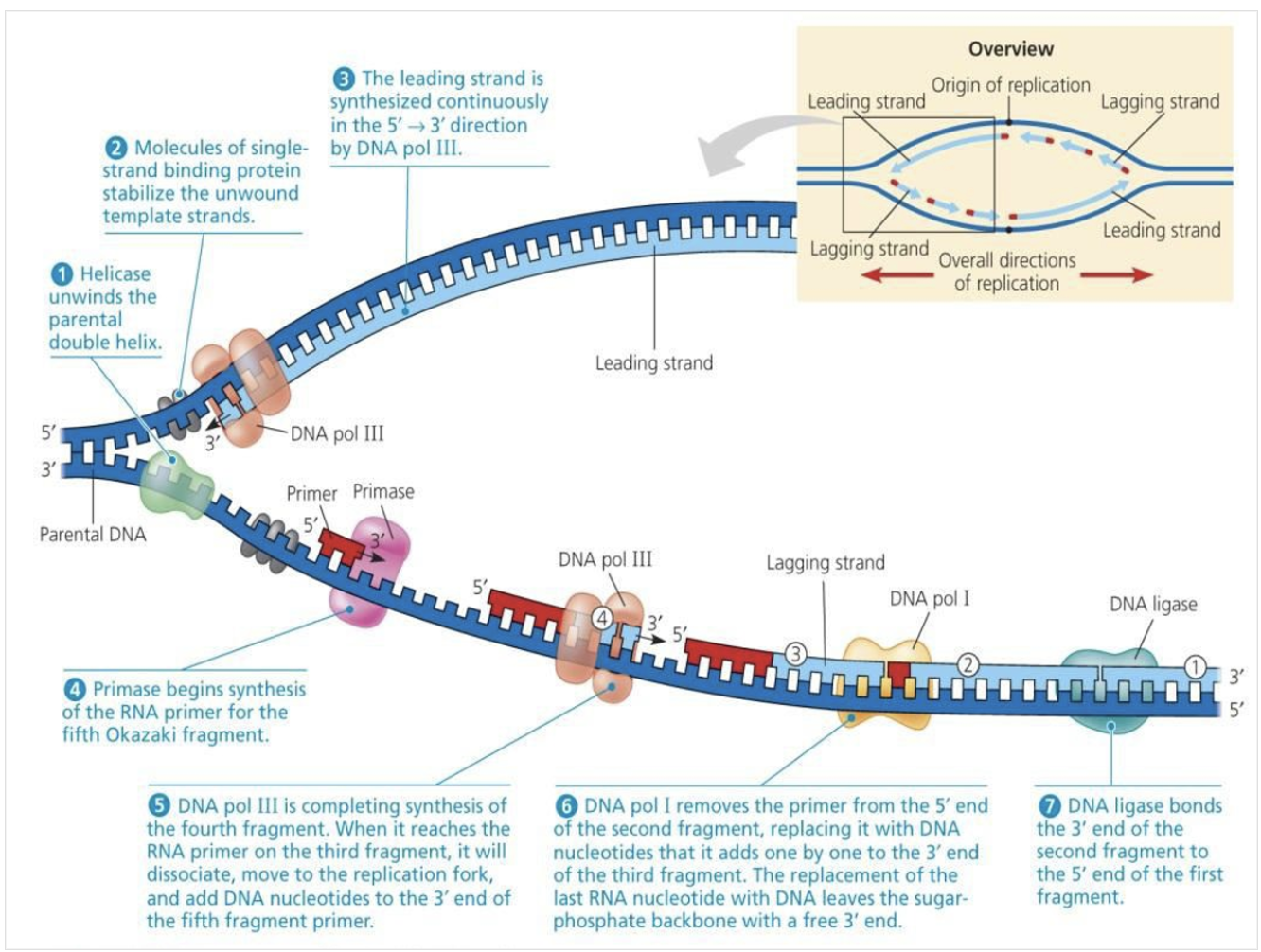
*After RNA primer is made, DNA pol III starts to synthesise the leading strand
*The leading strand/ continuous strand is elongated continuously in the 5->3 direction as the fork progresses
1) Topiomerase break, swivel, and rejoin (prevent supercoiling) ahead of helicase
2) Helicase cut the hydrogen bonding and open the DNA double strand
3) SSBP bind to each of the strand to prevent closing
4) When primase sythesize DNA primer, DNA polymerase3 comes to the 3' of the primer to elongate it.
5) When DNA polymerase3 reaches the 5' of the okazaki fragment ahead of it, it dissacociates
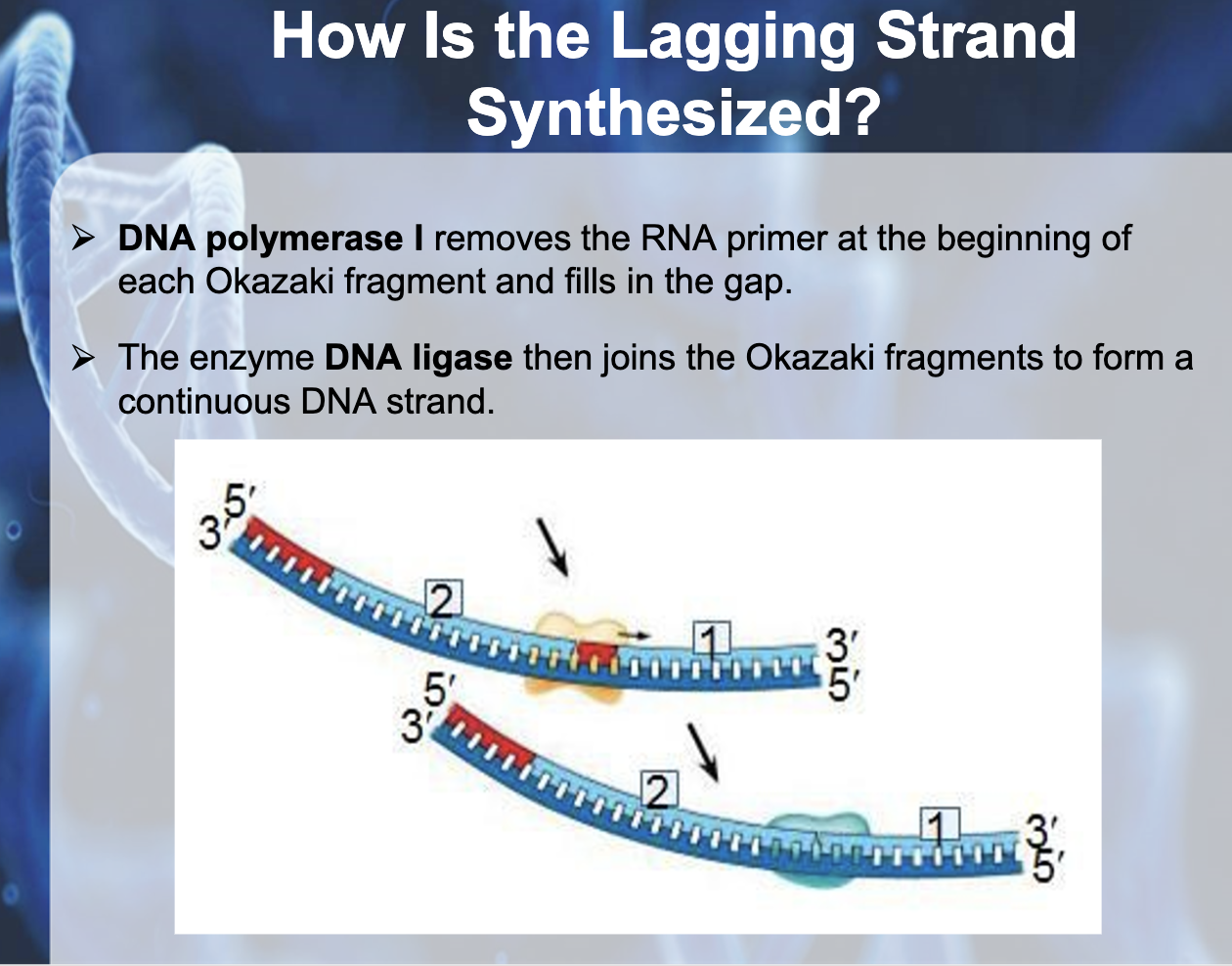
6) DNA polymerase1 replace RNA primer to DNA
7) Ligase glue to Okazaki fragments
DNA polymerase can only attach nucleotides to the 3' end of carbon, so the synthesis direction should always have to be 5'->3'
Lagging strand in turn can synthesise DNA uncontinuously.
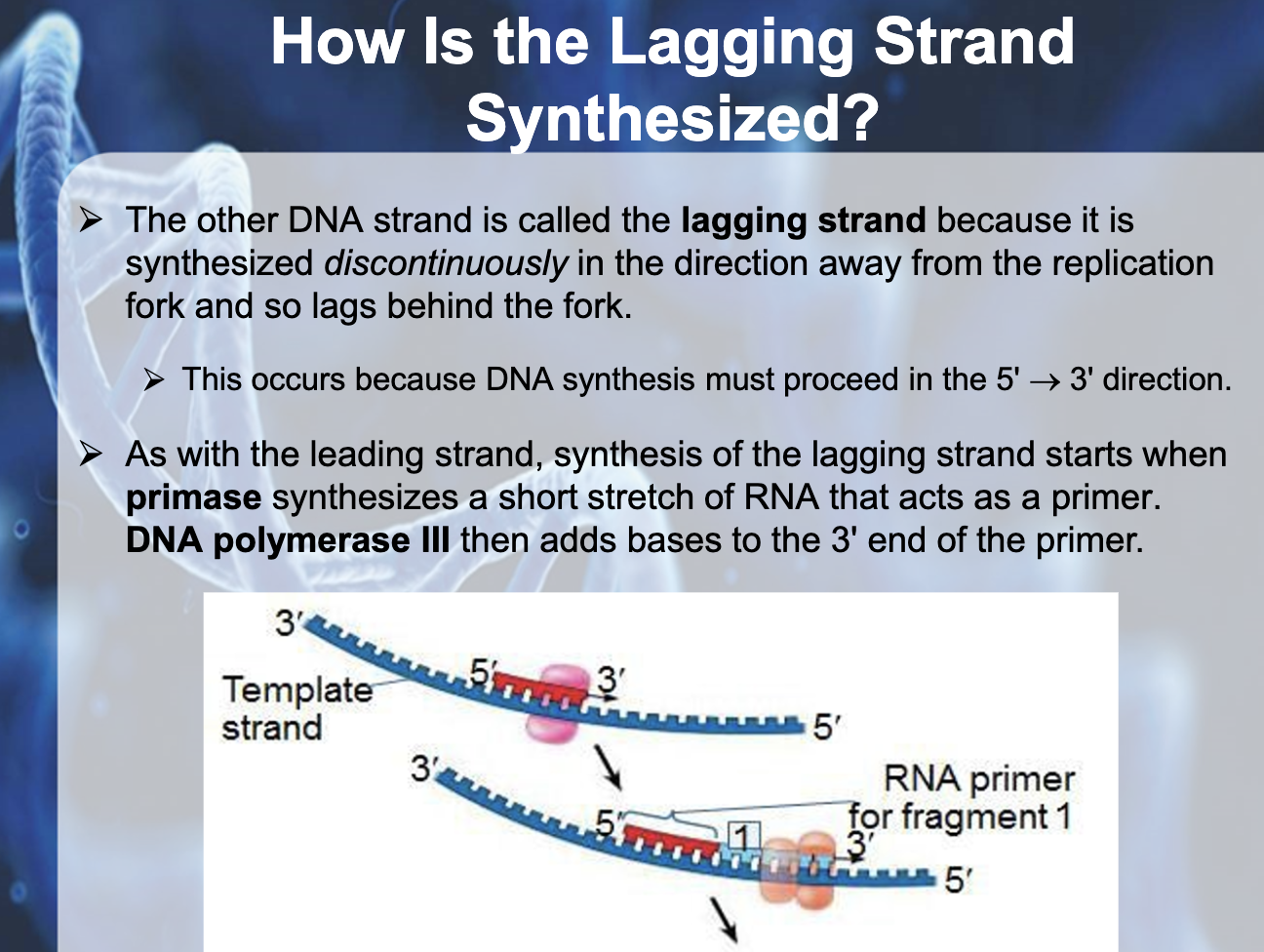
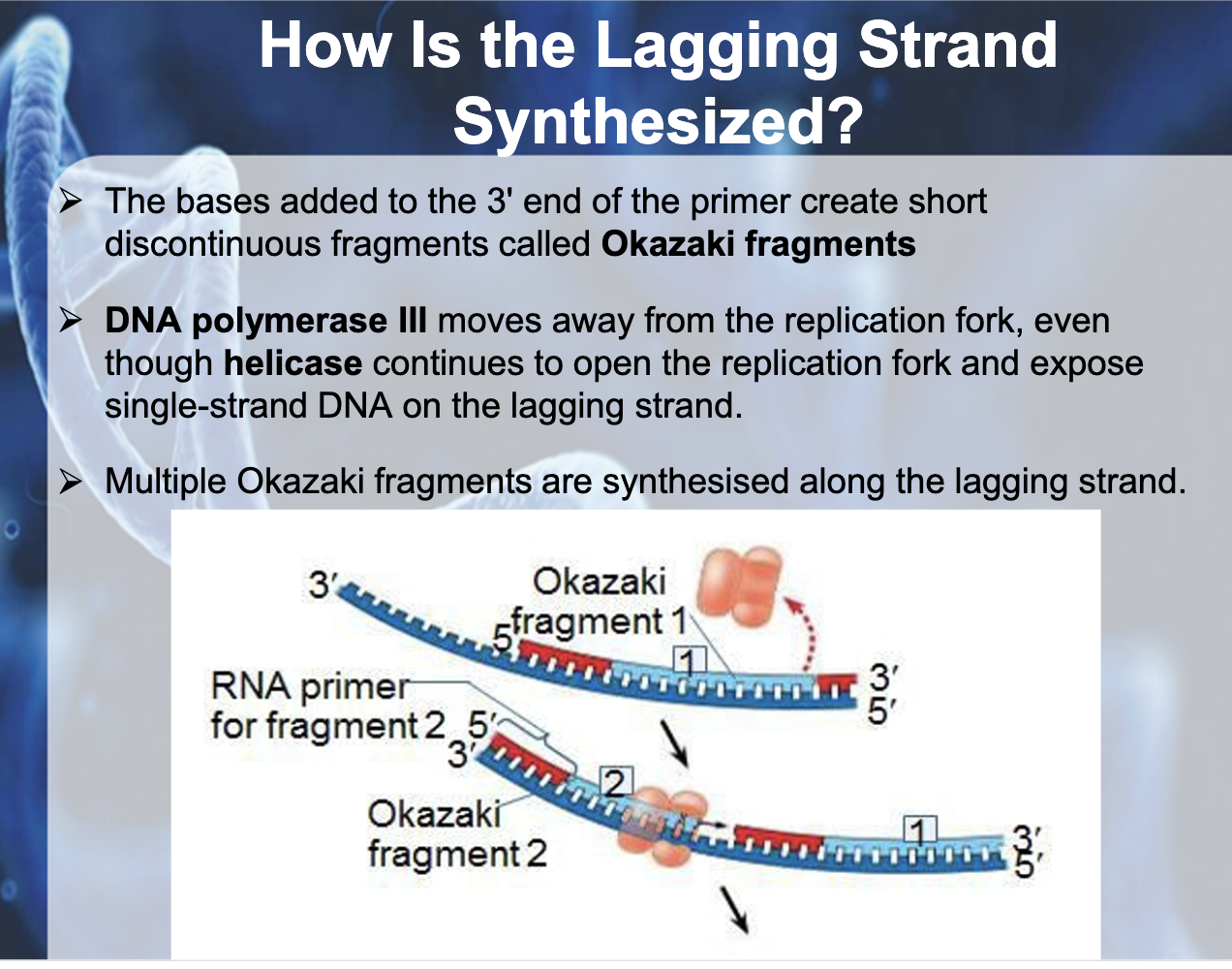
*Okazaki fragments : the short fragments created from the bases added to the 3' end of the primer
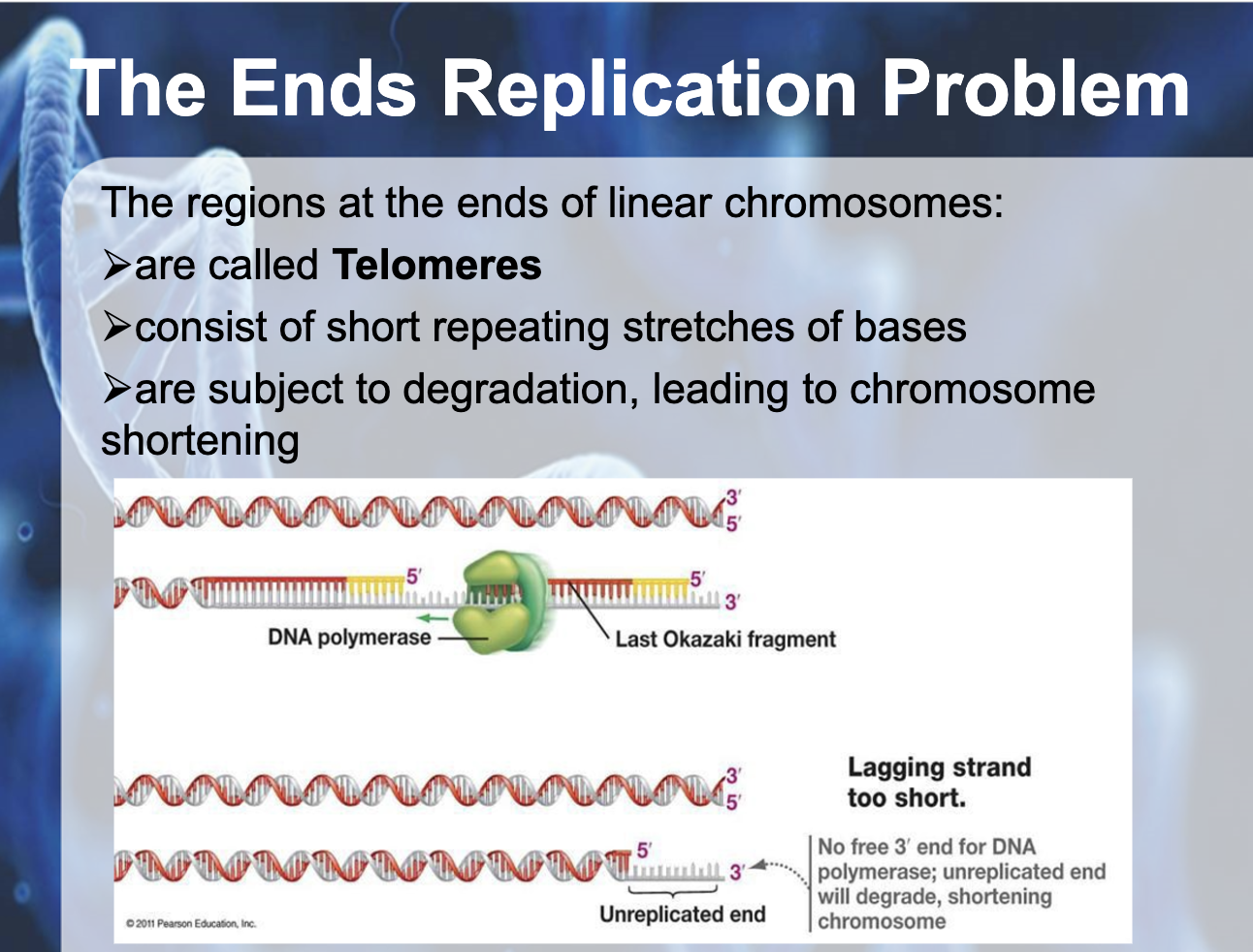
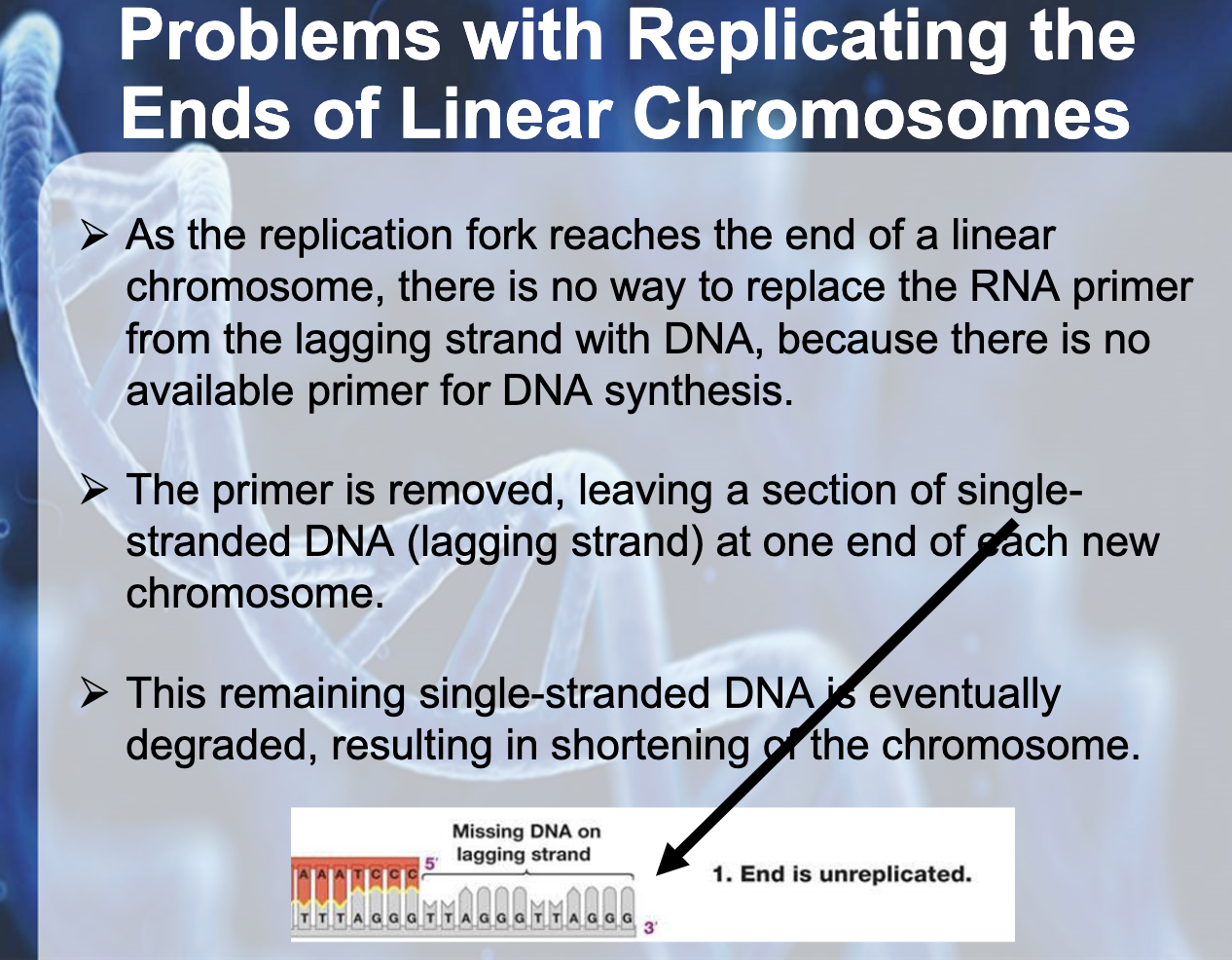
The end of the linear chromosome is called telomere
As there is no place for RNA primase to attach to make a primer (lagging strand) , telomerase adds more repeating bases to the end of the lagging strand (as a temporary template), catalyzing the synthesis of DNA from the telomerase template
-> Telmerase prevents the lagging strand from getting shorter with replication
(Somatic cells progressively shorten as the individual ages, they are lack of telomerase)
(Then Somatic cells' new DNA template from leading strand has length of as original state but only lagging strand shorten??)
(Telomere shortening has a role in limiting amount of time cells remaining in actively growing state)


https://www.youtube.com/watch?v=Qqe4thU-os8

REPLICATION MISTAKES

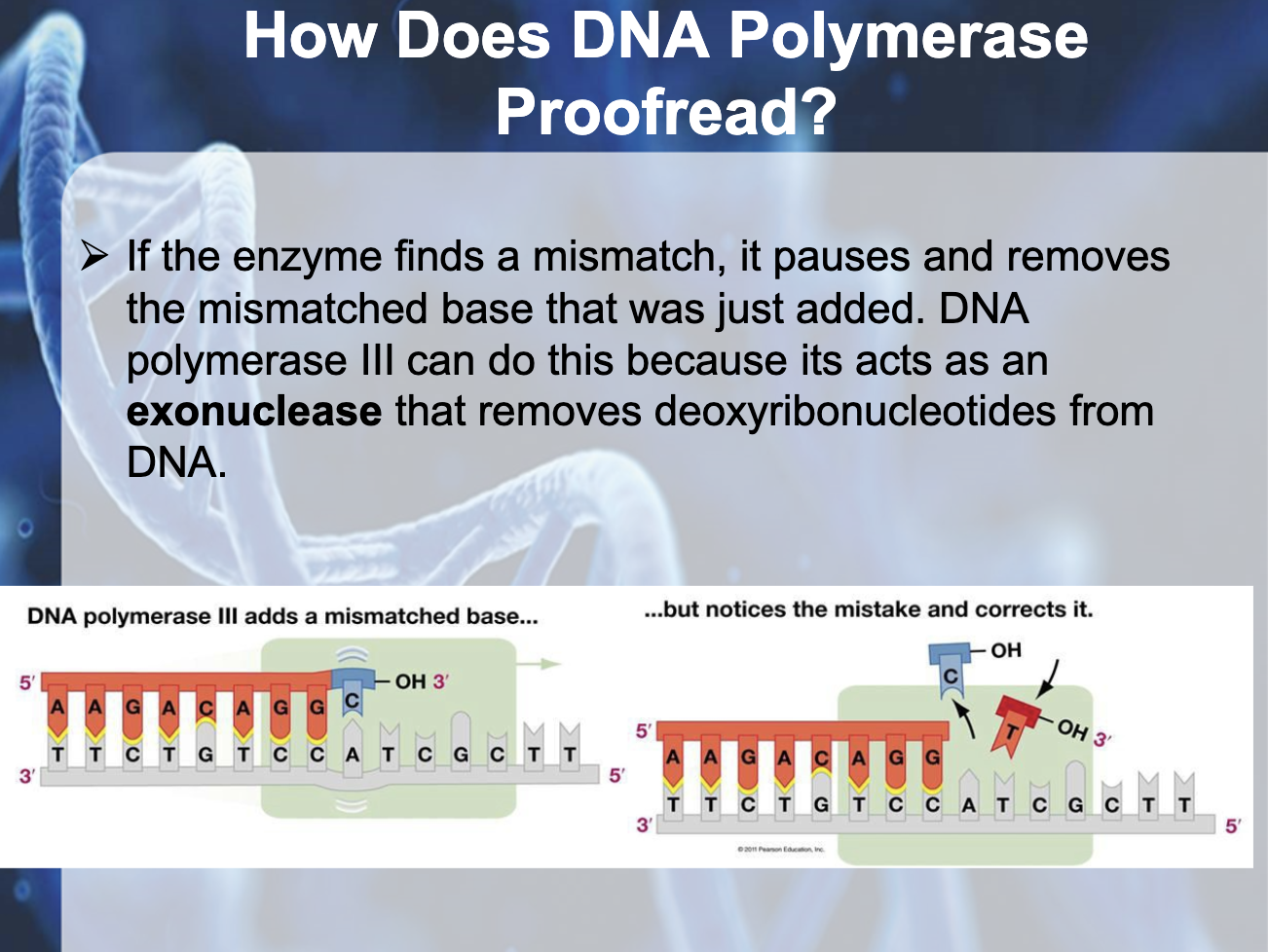
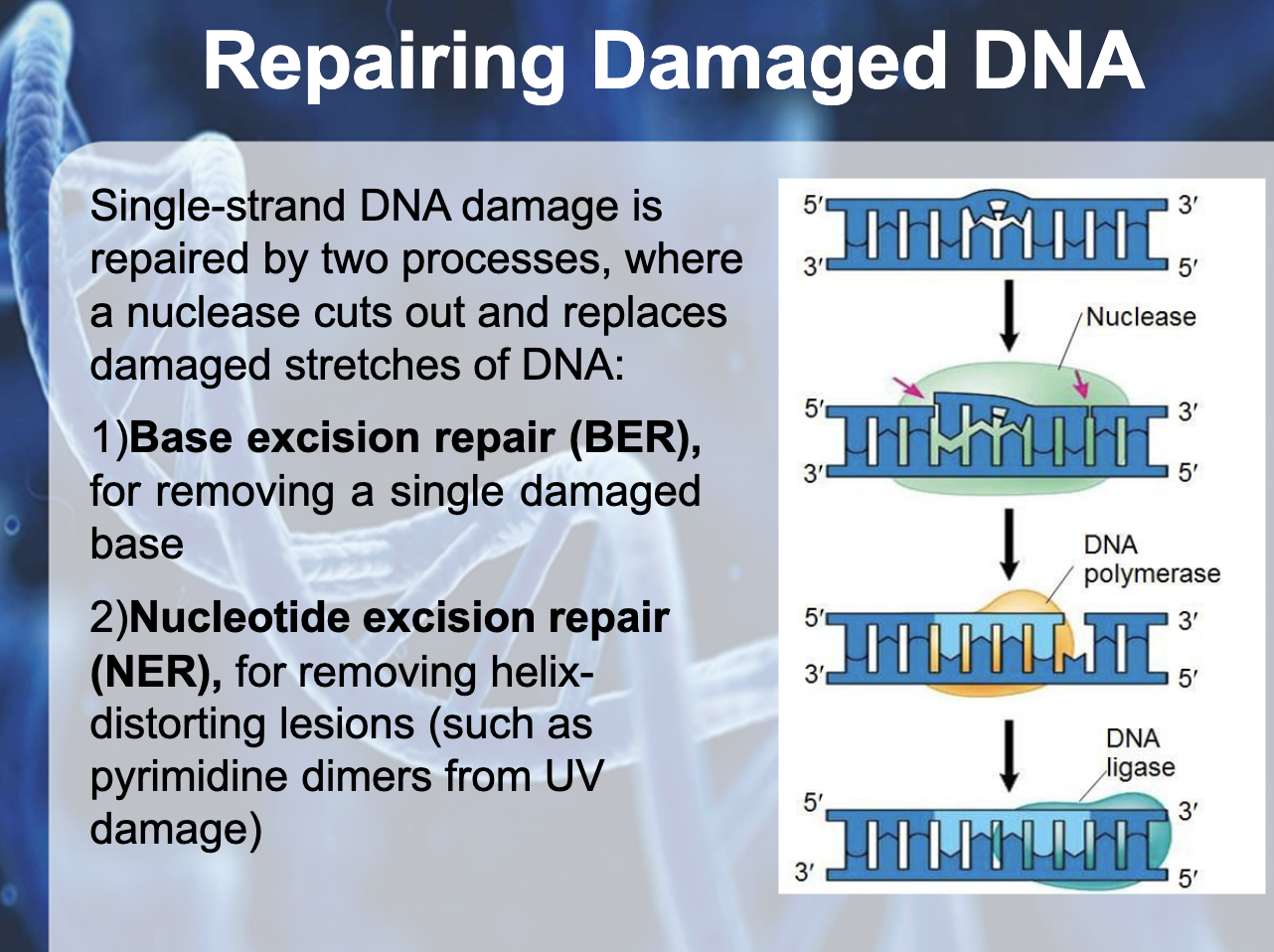
*Exonucleases are key enzymes involved in many aspects of cellular metabolism and maintenance and are essential to genome stability, acting to cleave DNA from free ends.
*Mismatch repair (MMR) relies upon the original strand (parental strand) to use as template for repairing the mismatch
*Single-strand DNA damage can also be caused by
-exposure to harmful chemical
-physical agents (cigarette, x-rays)
-spontaneous changes from cellular metabolic products (reactive oxygen species ROS)
1)Base excision repair (BER) : removes a single damaged base
2)Nucleotide excision repair (NER) : removes helix-distorting lesions
DNA PACKAGING

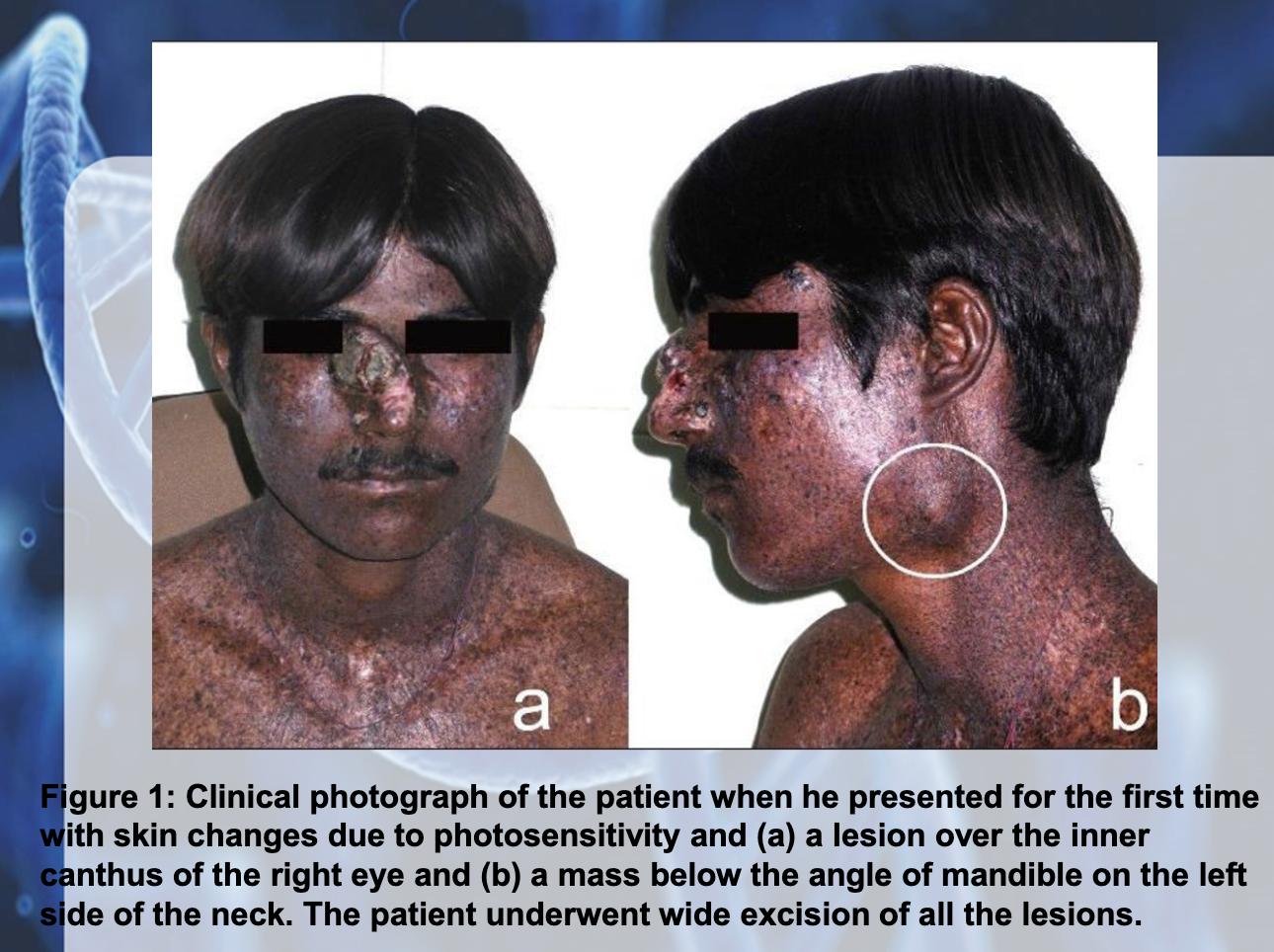
XERDOERMA PEGMENTOSUM (XP)
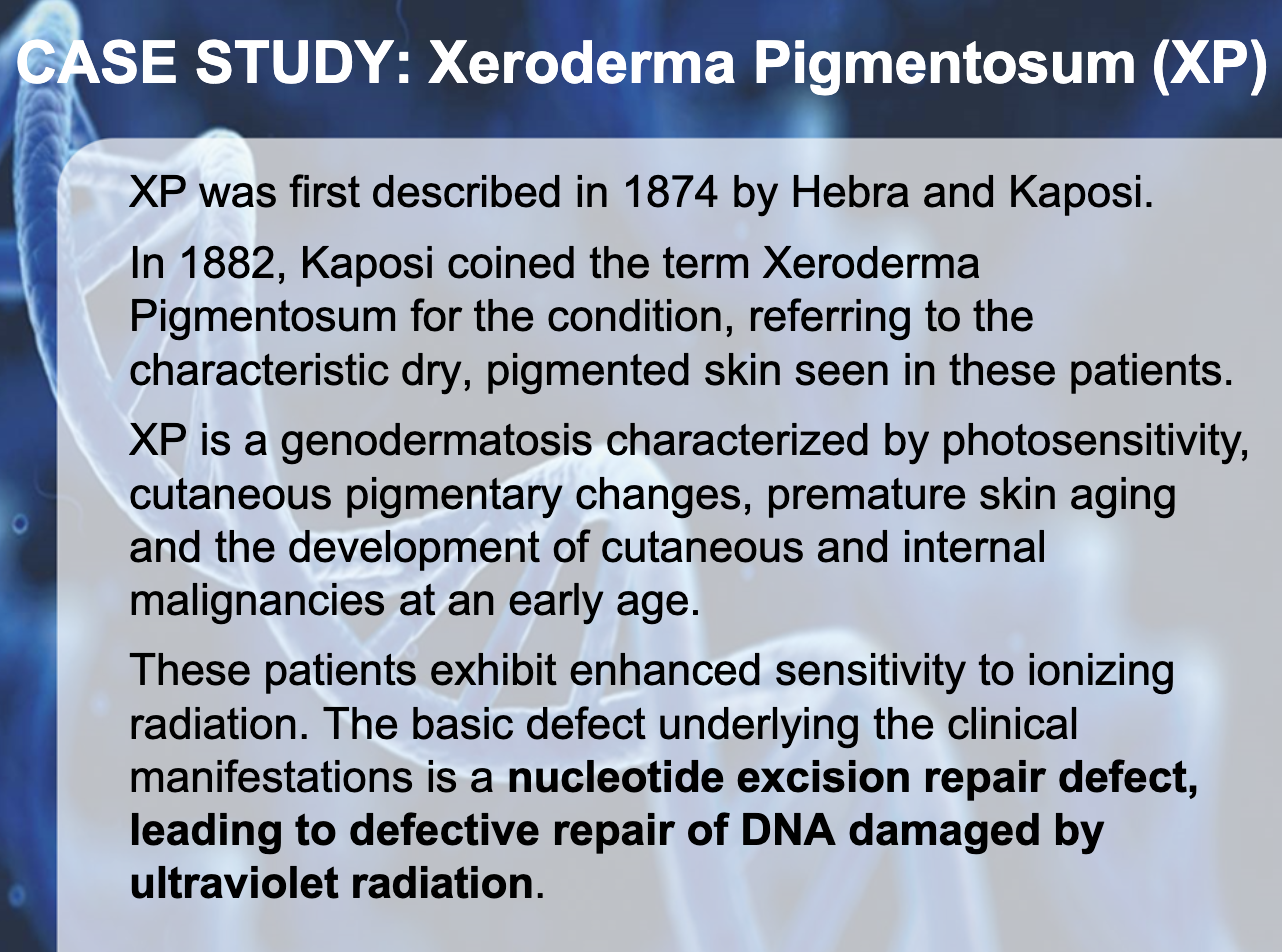
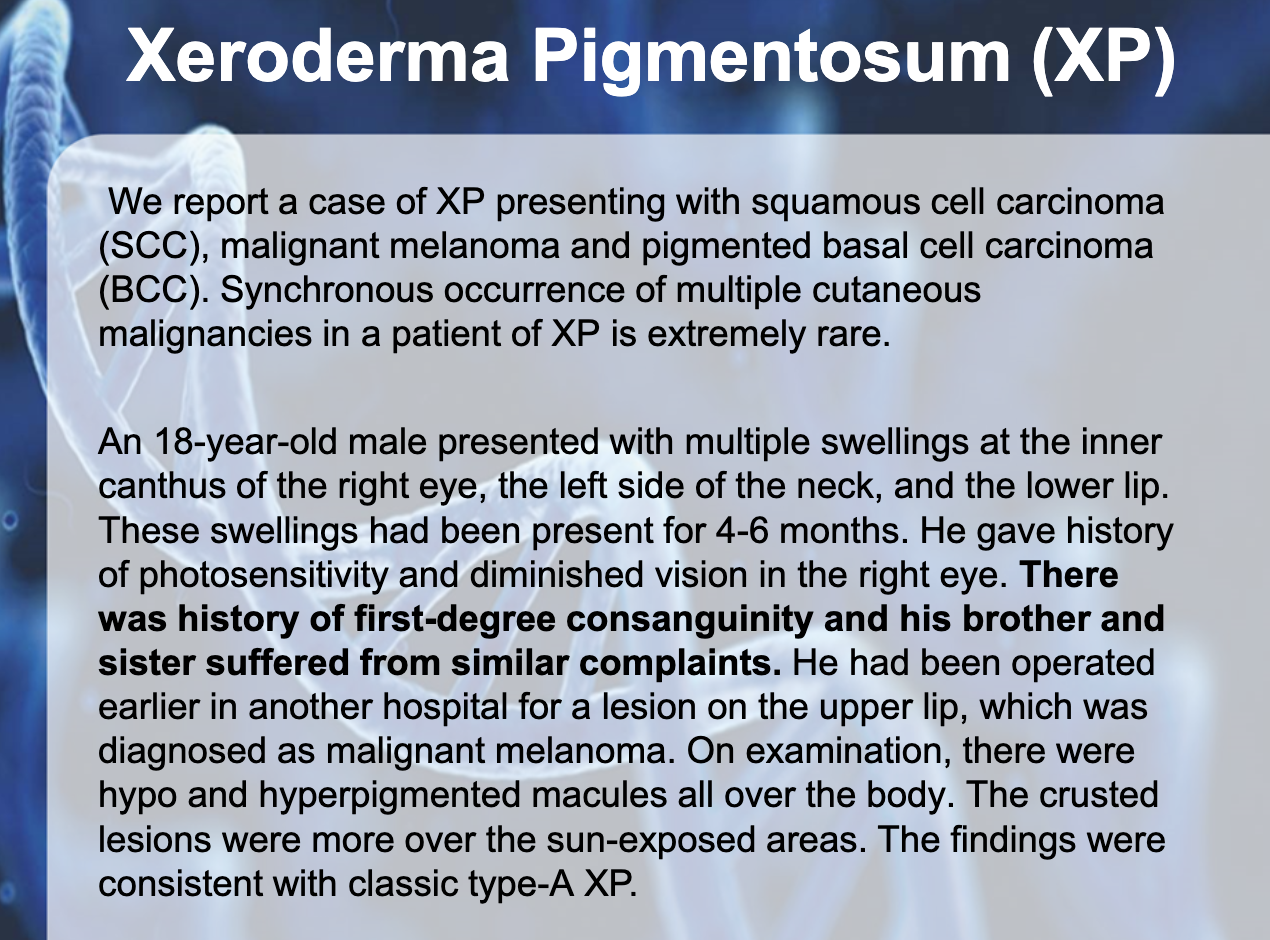
-Genodermatosis
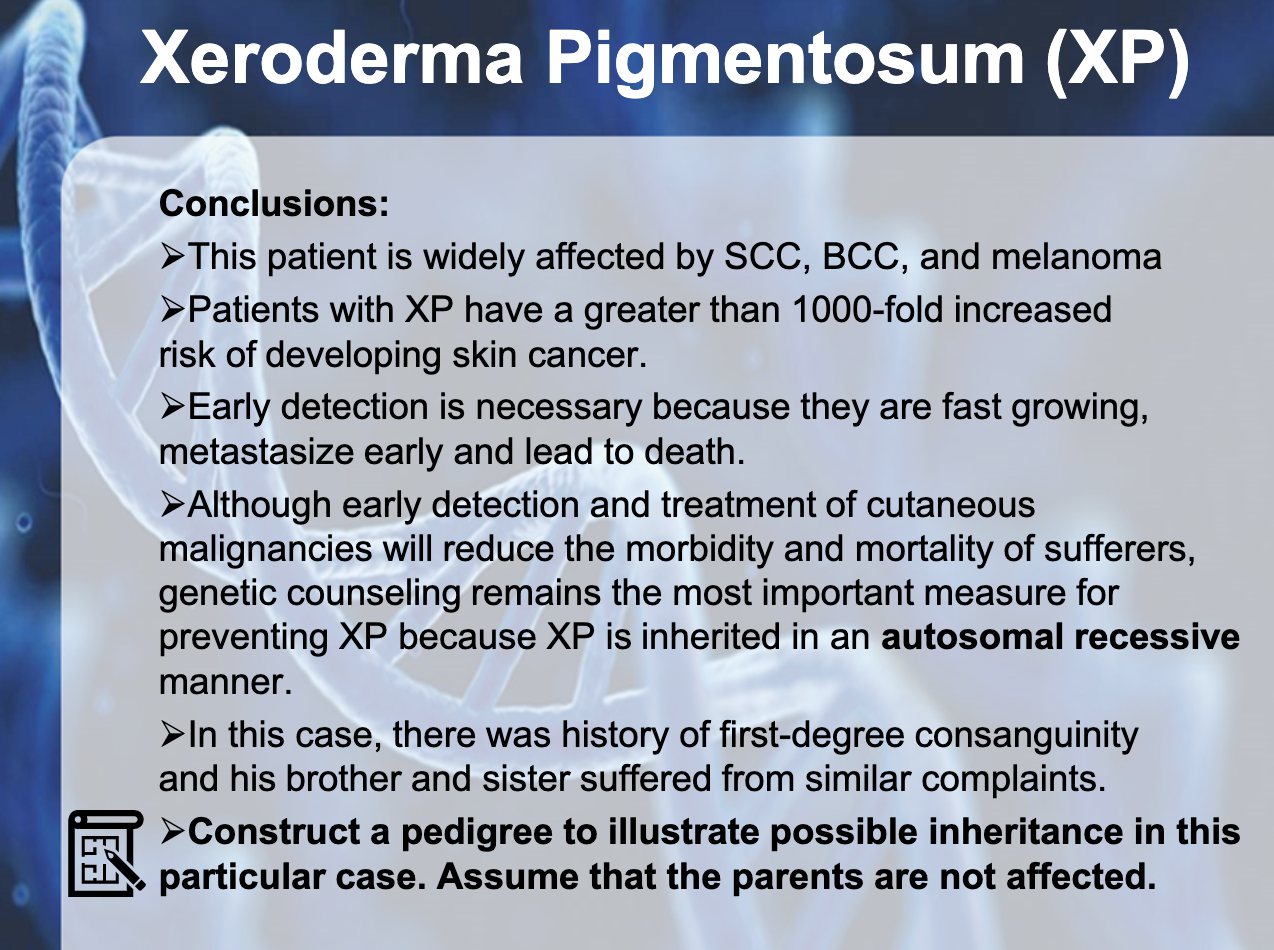
색소성 건피증(色素性乾皮症, xeroderma pigmentosum)은 DNA 수선의 희귀 상염색체 열성 유전병으로, 자외선에 의한 상해를 치료하는 능력이 결핍되는 질병이다.
SELF PACED QUIZZ

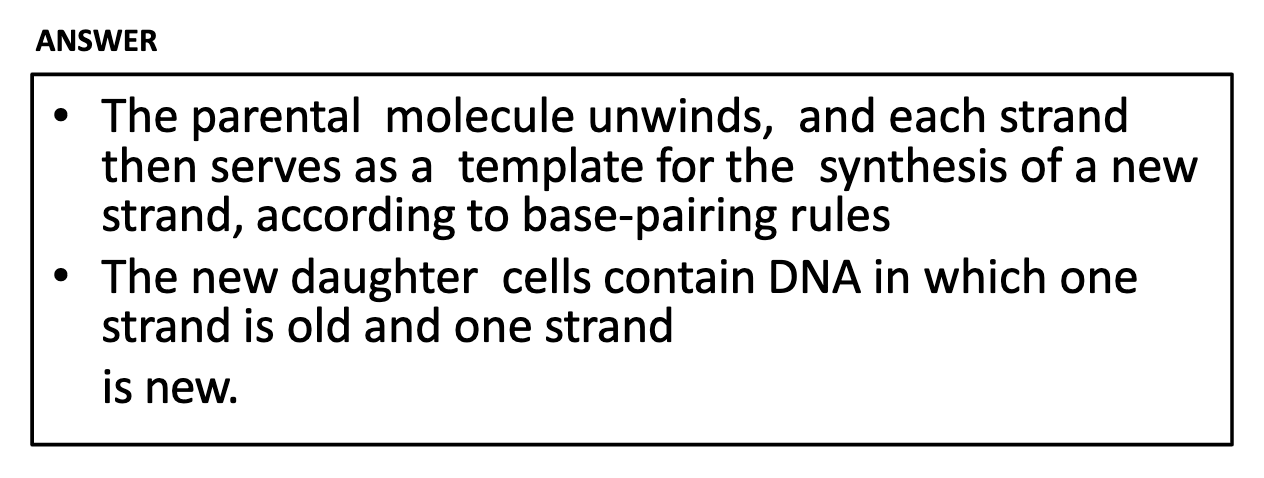
-Two strands of DNA are unwinded in replication and Each strand of DNA is used as a template for the synthesis of a new strand. As a result, the new daughter cells contain DNA where one strand is old and one strand is new
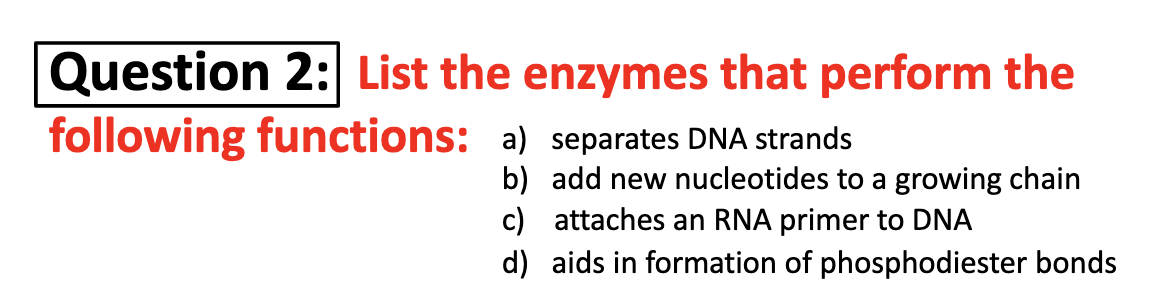

(a)Helicase (b)DNA polymerase (c)primase (d)ligase




'Griffith college Tri1 2023 > 1005 QBT (GnD)' 카테고리의 다른 글
| [WEEK8] Part 3 Developmental Genetics (0) | 2023.04.30 |
|---|---|
| [WEEK6] Part 1 Gene Expression (0) | 2023.04.12 |
| [WEEK4] Part 2 Chromosomal Basis of Inheritance (0) | 2023.04.01 |
| [WEEK3] Part 1 Mendelian Genetics (0) | 2023.03.16 |
| [WEEK2] Part2 Meiosis (0) | 2023.03.15 |



